Downregulation of the FTO m6A RNA demethylase promotes EMT-mediated progression of epithelial tumors and sensitivity to Wnt inhibitors
- PMID: 35121941
- PMCID: PMC10734094
- DOI: 10.1038/s43018-021-00223-7
Downregulation of the FTO m6A RNA demethylase promotes EMT-mediated progression of epithelial tumors and sensitivity to Wnt inhibitors
Abstract
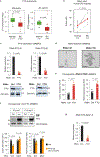
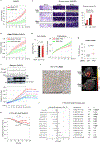
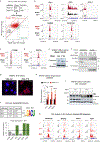
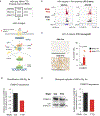
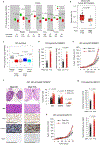
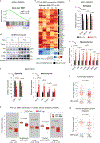
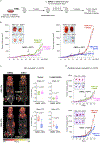
Post-transcriptional modifications of RNA constitute an emerging regulatory layer of gene expression. The demethylase fat mass- and obesity-associated protein (FTO), an eraser of N6-methyladenosine (m6A), has been shown to play a role in cancer, but its contribution to tumor progression and the underlying mechanisms remain unclear. Here, we report widespread FTO downregulation in epithelial cancers associated with increased invasion, metastasis and worse clinical outcome. Both in vitro and in vivo, FTO silencing promotes cancer growth, cell motility and invasion. In human-derived tumor xenografts (PDXs), FTO pharmacological inhibition favors tumorigenesis. Mechanistically, we demonstrate that FTO depletion elicits an epithelial-to-mesenchymal transition (EMT) program through increased m6A and altered 3'-end processing of key mRNAs along the Wnt signaling cascade. Accordingly, FTO knockdown acts via EMT to sensitize mouse xenografts to Wnt inhibition. We thus identify FTO as a key regulator, across epithelial cancers, of Wnt-triggered EMT and tumor progression and reveal a therapeutically exploitable vulnerability of FTO-low tumors.
© 2021. The Author(s), under exclusive licence to Springer Nature America, Inc.
Figures








Comment in
-
A Wnt twist in FTO's role in cancer progression.Nat Cancer. 2021 Jun;2(6):579-580. doi: 10.1038/s43018-021-00224-6. Nat Cancer. 2021. PMID: 35121942 No abstract available.
References
-
- Dominissini D. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 (2012). - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

