Small-molecule inhibitors that disrupt the MTDH-SND1 complex suppress breast cancer progression and metastasis
- PMID: 35121987
- PMCID: PMC8818087
- DOI: 10.1038/s43018-021-00279-5
Small-molecule inhibitors that disrupt the MTDH-SND1 complex suppress breast cancer progression and metastasis
Abstract
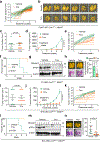
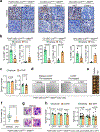
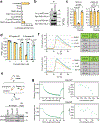
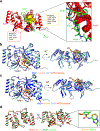
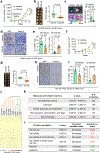
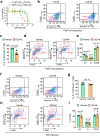
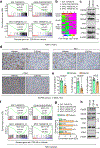
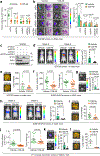
Metastatic breast cancer is a leading health burden worldwide. Previous studies have shown that metadherin (MTDH) promotes breast cancer initiation, metastasis and therapy resistance; however, the therapeutic potential of targeting MTDH remains largely unexplored. Here, we used genetically modified mice and demonstrate that genetic ablation of Mtdh inhibits breast cancer development through disrupting the interaction with staphylococcal nuclease domain-containing 1 (SND1), which is required to sustain breast cancer progression in established tumors. We performed a small-molecule compound screening to identify a class of specific inhibitors that disrupts the protein-protein interaction (PPI) between MTDH and SND1 and show that our lead candidate compounds C26-A2 and C26-A6 suppressed tumor growth and metastasis and enhanced chemotherapy sensitivity in preclinical models of triple-negative breast cancer (TNBC). Our results demonstrate a significant therapeutic potential in targeting the MTDH-SND1 complex and identify a new class of therapeutic agents for metastatic breast cancer.
© 2021. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests
Princeton has filed a disclosure on the findings based on this study. Y.K., M.S. and H.K. are named as co-inventors on the disclosure. J.F.J. is a co-founder and Y.K. is a co-founder and chair of scientific advisory board of Firebrand Therapeutics Inc., which has licensed relevant technologies from Princeton University to develop MTDH-SND1 targeting therapeutics. Y.K. is also a co-founder of Kayothera, Inc. and a member of Scientific Advisory Board for Cytocare, Inc and Vibrant Pharma Limited. M.R. is an employee of Crelux GmgH. S.R. is the owner of SWR Pharma Consulting LLC. The remaining authors declare no competing interests.
Figures









Comment in
-
Breaking up MTDH-SND1 to break down metastasis.Nat Cancer. 2022 Jan;3(1):6-8. doi: 10.1038/s43018-021-00320-7. Nat Cancer. 2022. PMID: 35121995 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

