Evolutionary history of type II transmembrane serine proteases involved in viral priming
- PMID: 35122525
- PMCID: PMC8817155
- DOI: 10.1007/s00439-022-02435-y
Evolutionary history of type II transmembrane serine proteases involved in viral priming
Abstract
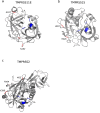
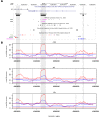
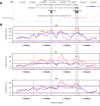
Type II transmembrane serine proteases (TTSPs) are a family of trypsin-like membrane-anchored serine proteases that play key roles in the regulation of some crucial processes in physiological conditions, including cardiac function, digestion, cellular iron homeostasis, epidermal differentiation, and immune responses. However, some of them, in particular TTSPs expressed in the human airways, were identified as host factors that promote the proteolytic activation and spread of respiratory viruses such as influenza virus, human metapneumovirus, and coronaviruses, including SARS-CoV-2. Given their involvement in viral priming, we hypothesized that members of the TTSP family may represent targets of positive selection, possibly as the result of virus-driven pressure. Thus, we investigated the evolutionary history of sixteen TTSP genes in mammals. Evolutionary analyses indicate that most of the TTSP genes that have a verified role in viral proteolytic activation present signals of pervasive positive selection, suggesting that viral infections represent a selective pressure driving the evolution of these proteases. We also evaluated genetic diversity in human populations and we identified targets of balancing selection in TMPRSS2 and TMPRSS4. This scenario may be the result of an ancestral and still ongoing host-pathogen arms race. Overall, our results provide evolutionary information about candidate functional sites and polymorphic positions in TTSP genes.
© 2022. The Author(s), under exclusive licence to Springer-Verlag GmbH Germany, part of Springer Nature.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

