FastSurferVINN: Building resolution-independence into deep learning segmentation methods-A solution for HighRes brain MRI
- PMID: 35122967
- PMCID: PMC9801435
- DOI: 10.1016/j.neuroimage.2022.118933
FastSurferVINN: Building resolution-independence into deep learning segmentation methods-A solution for HighRes brain MRI
Abstract
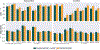
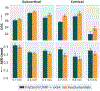
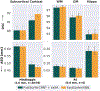
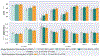
Leading neuroimaging studies have pushed 3T MRI acquisition resolutions below 1.0 mm for improved structure definition and morphometry. Yet, only few, time-intensive automated image analysis pipelines have been validated for high-resolution (HiRes) settings. Efficient deep learning approaches, on the other hand, rarely support more than one fixed resolution (usually 1.0 mm). Furthermore, the lack of a standard submillimeter resolution as well as limited availability of diverse HiRes data with sufficient coverage of scanner, age, diseases, or genetic variance poses additional, unsolved challenges for training HiRes networks. Incorporating resolution-independence into deep learning-based segmentation, i.e., the ability to segment images at their native resolution across a range of different voxel sizes, promises to overcome these challenges, yet no such approach currently exists. We now fill this gap by introducing a Voxel-size Independent Neural Network (VINN) for resolution-independent segmentation tasks and present FastSurferVINN, which (i) establishes and implements resolution-independence for deep learning as the first method simultaneously supporting 0.7-1.0 mm whole brain segmentation, (ii) significantly outperforms state-of-the-art methods across resolutions, and (iii) mitigates the data imbalance problem present in HiRes datasets. Overall, internal resolution-independence mutually benefits both HiRes and 1.0 mm MRI segmentation. With our rigorously validated FastSurferVINN we distribute a rapid tool for morphometric neuroimage analysis. The VINN architecture, furthermore, represents an efficient resolution-independent segmentation method for wider application.
Keywords: Artificial intelligence; Computational neuroimaging; Deep learning; High-resolution; Structural MRI.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures




References
-
- A mind-brain, 2019. -Body dataset of MRI, EEG, cognition, emotion, and peripheral physiology in young and old adults. Sci. Data 6 (1), 180308. doi: 10.1038/sdata.2018.308. http://www.nature.com/articles/sdata2018308 - DOI - PMC - PubMed
-
- Alao H, Kim J-S, Kim TS, Lee K, 2021. Efficient multi-scalable network for single image super resolution. J. Multimed. Inf. Syst 8 (2), 101–110. doi: 10.33851/JMIS.2021.8.2.101. - DOI
-
- Allebach JP, 2005. 7.1 - image scanning, sampling, and interpolation. In: Bovik A (Ed.). Communications, Networking and Multimedia, Handbook of Image and Video Processing, second ed., Academic Press, Burlington: doi: 10.1016/B978-012119792-6/50115-7. pp. 895–XXVII. https://www.sciencedirect.com/science/article/pii/B9780121197926501157 - DOI
-
- Benjamini Y, Hochberg Y, 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc 57 (1), 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. https://rss.onlinelibrary.wiley.com/doi/pdf/10.1111/j.2517-6161.1995.tb0... - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

