Development and Validation of Two RT-qPCR Diagnostic Assays for Detecting Severe Acute Respiratory Syndrome Coronavirus 2 Genomic Targets across Two Specimen Types
- PMID: 35124239
- PMCID: PMC8813189
- DOI: 10.1016/j.jmoldx.2021.12.010
Development and Validation of Two RT-qPCR Diagnostic Assays for Detecting Severe Acute Respiratory Syndrome Coronavirus 2 Genomic Targets across Two Specimen Types
Abstract
Following the outbreak and subsequent pandemic of coronavirus disease 2019 (COVID-19), clinical diagnostic laboratories worldwide sought accurate and reliable testing methodologies. However, many laboratories were and still are hindered by a number of factors, including an unprecedented demand for testing, reagent and laboratory supply shortages and availability of qualified staff. To respond to these concerns, two separate laboratory-developed tests were validated for detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) using two different specimen types. In addition, these assays target different genomic regions of SARS-CoV-2, allowing for viral detection and mitigating genetic variation. Lower limit of detection and clinical evaluation studies showed detection of SARS-CoV-2 at 500 cp/mL with nasopharyngeal and saliva samples. These multiplexed RT-qPCR assays, although based on modified CDC, New York State Department of Health, and World Health Organization Emergency Use Authorization tests, allow for higher throughput and rapid turnaround time, benefiting patients, clinicians, and communities as a whole. These cost-effective tests also use readily obtainable reagents, circumventing commercial assay supply chain issues. The laboratory-developed tests described here have improved patient care and are highly adaptable should the need arise at other clinical diagnostic laboratories. Furthermore, the foundation and design of these assays may be modified in the future for detection of COVID-19 variants or other RNA-based viral detection tests.
Copyright © 2022 Association for Molecular Pathology and American Society for Investigative Pathology. Published by Elsevier Inc. All rights reserved.
Figures
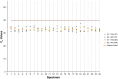
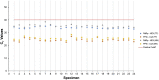
References
-
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W., China Novel Coronavirus Investigating and Research Team A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. - PMC - PubMed
-
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probably bat origin. Nature. 2020;579:270–273. - PMC - PubMed
-
- Finkel Y., Mizrahi O., Nachshon A., Weingarten-Gabbay S., Morgenstern D., Yahalom-Ronen Y., Tamir H., Achdout H., Stein D., Israeli O., Beth-Din A., Melamed S., Weiss S., Israely T., Paran N., Schwartz M., Stern-Ginossar N. The coding capacity of SARS-CoV-2. Nature. 2021;589:125–130. - PubMed
-
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

