Identification of an Aptamer With Binding Specificity to Tumor-Homing Myeloid-Derived Suppressor Cells
- PMID: 35126104
- PMCID: PMC8814529
- DOI: 10.3389/fphar.2021.752934
Identification of an Aptamer With Binding Specificity to Tumor-Homing Myeloid-Derived Suppressor Cells
Abstract
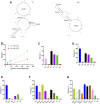
Myeloid-derived suppressor cells (MDSCs) play a critical role in tumor growth and metastasis. Since they constantly infiltrate into the tumor tissue, these cells are considered as an ideal carrier for tumor-targeted drug delivery. We recently identified a DNA-based thioaptamer (T1) with tumor accumulating activity, demonstrated its potential on tumor targeting and drug delivery. In the current study, we have carried out structure-activity relationship analysis to further optimize the aptamer. In the process, we have identified a sequence-modified aptamer (M1) that shows an enhanced binding affinity to MDSCs over the parental T1 aptamer. In addition, M1 can penetrate into the tumor tissue more effectively by hitchhiking on MDSCs. Taken together, we have identified a new reagent for enhanced tumor-targeted drug delivery.
Keywords: G-quadruplex; aptamer; myeloid-derived suppressor cell; structure-activity relationship; tumor-targeted delivery.
Copyright © 2022 Tian, Welte, Mai, Liu, Ramirez and Shen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Azri F. A., Selamat J., Sukor R., Yusof N. A., Raston N. H. A., Eissa S., et al. (2021). Determination of Minimal Sequence for Zearalenone Aptamer by Computational Docking and Application on an Indirect Competitive Electrochemical Aptasensor. Anal. Bioanal. Chem. 413 (15), 3861–3872. 10.1007/s00216-021-03336-1 - DOI - PubMed
-
- Bosiljcic M., Cederberg R. A., Hamilton M. J., LePard N. E., Harbourne B. T., Collier J. L., et al. (2019). Targeting Myeloid-Derived Suppressor Cells in Combination with Primary Mammary Tumor Resection Reduces Metastatic Growth in the Lungs. Breast Cancer Res. 21 (1), 103. 10.1186/s13058-019-1189-x - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

