DDIT: An Online Predictor for Multiple Clinical Phenotypic Drug-Disease Associations
- PMID: 35126114
- PMCID: PMC8809407
- DOI: 10.3389/fphar.2021.772026
DDIT: An Online Predictor for Multiple Clinical Phenotypic Drug-Disease Associations
Abstract
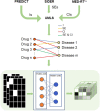
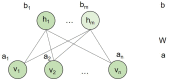
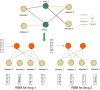
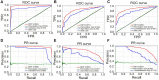
Background: Drug repurposing provides an effective method for high-speed, low-risk drug development. Clinical phenotype-based screening exceeded target-based approaches in discovering first-in-class small-molecule drugs. However, most of these approaches predict only binary phenotypic associations between drugs and diseases; the types of drug and diseases have not been well exploited. Principally, the clinical phenotypes of a known drug can be divided into indications (Is), side effects (SEs), and contraindications (CIs). Incorporating these different clinical phenotypes of drug-disease associations (DDAs) can improve the prediction accuracy of the DDAs. Methods: We develop Drug Disease Interaction Type (DDIT), a user-friendly online predictor that supports drug repositioning by submitting known Is, SEs, and CIs for a target drug of interest. The dataset for Is, SEs, and CIs was extracted from PREDICT, SIDER, and MED-RT, respectively. To unify the names of the drugs and diseases, we mapped their names to the Unified Medical Language System (UMLS) ontology using Rest API. We then integrated multiple clinical phenotypes into a conditional restricted Boltzmann machine (RBM) enabling the identification of different phenotypes of drug-disease associations, including the prediction of as yet unknown DDAs in the input. Results: By 10-fold cross-validation, we demonstrate that DDIT can effectively capture the latent features of the drug-disease association network and represents over 0.217 and over 0.072 improvement in AUC and AUPR, respectively, for predicting the clinical phenotypes of DDAs compared with the classic K-nearest neighbors method (KNN, including drug-based KNN and disease-based KNN), Random Forest, and XGBoost. By conducting leave-one-drug-class-out cross-validation, the AUC and AUPR of DDIT demonstrated an improvement of 0.135 in AUC and 0.075 in AUPR compared to any of the other four methods. Within the top 10 predicted indications, side effects, and contraindications, 7/10, 9/10, and 9/10 hit known drug-disease associations. Overall, DDIT is a useful tool for predicting multiple clinical phenotypic types of drug-disease associations.
Keywords: contraindication; drug repositioning; indication; machine learning; phenotypic types of drug-disease associations; restricted Boltzmann machine; side effect.
Copyright © 2022 Lu, Qin, Chen, Wu, Zhao, Miyano, Zhang, Yu and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
RLFDDA: a meta-path based graph representation learning model for drug-disease association prediction.BMC Bioinformatics. 2022 Dec 1;23(1):516. doi: 10.1186/s12859-022-05069-z. BMC Bioinformatics. 2022. PMID: 36456957 Free PMC article.
-
Neighborhood-based inference and restricted Boltzmann machine for microbe and drug associations prediction.PeerJ. 2022 Aug 15;10:e13848. doi: 10.7717/peerj.13848. eCollection 2022. PeerJ. 2022. PMID: 35990901 Free PMC article.
-
Drug repositioning of herbal compounds via a machine-learning approach.BMC Bioinformatics. 2019 May 29;20(Suppl 10):247. doi: 10.1186/s12859-019-2811-8. BMC Bioinformatics. 2019. PMID: 31138103 Free PMC article.
-
Predicting drug-target interactions using restricted Boltzmann machines.Bioinformatics. 2013 Jul 1;29(13):i126-34. doi: 10.1093/bioinformatics/btt234. Bioinformatics. 2013. PMID: 23812976 Free PMC article.
-
An effective drug-disease associations prediction model based on graphic representation learning over multi-biomolecular network.BMC Bioinformatics. 2022 Jan 4;23(1):9. doi: 10.1186/s12859-021-04553-2. BMC Bioinformatics. 2022. PMID: 34983364 Free PMC article.
References
LinkOut - more resources
Full Text Sources

