Lactic Acid Bacteria Are Prevalent in the Infrabuccal Pockets and Crops of Ants That Prefer Aphid Honeydew
- PMID: 35126329
- PMCID: PMC8814368
- DOI: 10.3389/fmicb.2021.785016
Lactic Acid Bacteria Are Prevalent in the Infrabuccal Pockets and Crops of Ants That Prefer Aphid Honeydew
Abstract
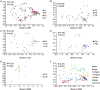
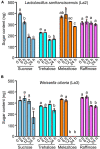
Ants are evolutionarily successful species and occupy diverse trophic and habitat niches on the earth. To fulfill dietary requirements, ants have established commensalism with both sap-feeding insects and bacteria. In this study, we used high-throughput sequencing of the bacterial 16S rRNA gene to characterize the bacterial composition and structure of the digestive tracts in three species of Formica ants and Lasius niger (Linnaeus)-species that predominantly feed on honeydew secreted by aphids. We found that bacterial communities displayed species- and colony-level signatures, and that bacterial communities in the infrabuccal pockets and crops were different from those in the midguts and hindguts. Lactobacillus and Wolbachia were dominant in the infrabuccal pockets and crops of workers, whereas Wolbachia was dominant in the midguts, hindguts and brood (larvae, pupae and cocoons). To learn more about the dominant Lactobacillus in ants, we assessed its prevalence in a wide range of aphid-tending ants using diagnostic PCR. We found that Lactobacillus was more prevalent in Formicinae than in Myrmicinae species. We also isolated four strains of lactic acid bacteria (Lactobacillus sanfranciscensis, Lactobacillus lindneri, Weissella cibaria and Fructobacillus sp.) from the infrabuccal pockets and crops of aphid-tending ants using a culture-dependent method. Two predominant lactic acid bacterial isolates, Lactobacillus sanfranciscensis (La2) and Weissella cibaria (La3), exhibited abilities in catabolizing sugars (sucrose, trehalose, melezitose and raffinose) known to be constituents of hemipteran honeydew. These findings contribute to further understanding the association between ants, aphids and bacteria, and provide additional information on the function of lactic acid bacteria in ants.
Keywords: ants; aphids; bacterial communities; digestive tract; lactic acid bacteria.
Copyright © 2022 Zheng, Zhao, Zhang, Hu, Xu, Wei and He.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Archie E. A., Theis K. R. (2011). Animal behaviour meets microbial ecology. Anim. Behav. 82 425–436. 10.1016/j.anbehav.2011.05.029 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

