Microbial Communities Influence Soil Dissolved Organic Carbon Concentration by Altering Metabolite Composition
- PMID: 35126334
- PMCID: PMC8811196
- DOI: 10.3389/fmicb.2021.799014
Microbial Communities Influence Soil Dissolved Organic Carbon Concentration by Altering Metabolite Composition
Abstract
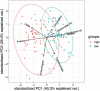
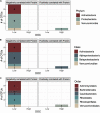
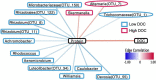
Rapid microbial growth in the early phase of plant litter decomposition is viewed as an important component of soil organic matter (SOM) formation. However, the microbial taxa and chemical substrates that correlate with carbon storage are not well resolved. The complexity of microbial communities and diverse substrate chemistries that occur in natural soils make it difficult to identify links between community membership and decomposition processes in the soil environment. To identify potential relationships between microbes, soil organic matter, and their impact on carbon storage, we used sand microcosms to control for external environmental factors such as changes in temperature and moisture as well as the variability in available carbon that exist in soil cores. Using Fourier transform ion cyclotron resonance mass spectrometry (FTICR-MS) on microcosm samples from early phase litter decomposition, we found that protein- and tannin-like compounds exhibited the strongest correlation to dissolved organic carbon (DOC) concentration. Proteins correlated positively with DOC concentration, while tannins correlated negatively with DOC. Through random forest, neural network, and indicator species analyses, we identified 42 bacterial and 9 fungal taxa associated with DOC concentration. The majority of bacterial taxa (26 out of 42 taxa) belonged to the phylum Proteobacteria while all fungal taxa belonged to the phylum Ascomycota. Additionally, we identified significant connections between microorganisms and protein-like compounds and found that most taxa (12/14) correlated negatively with proteins indicating that microbial consumption of proteins is likely a significant driver of DOC concentration. This research links DOC concentration with microbial production and/or decomposition of specific metabolites to improve our understanding of microbial metabolism and carbon persistence.
Keywords: DOC; FTICR mass spectrometry; bacteria; fungi; metabolites; microbial communities.
Copyright © 2022 Campbell, Ulrich, Toyoda, Thompson, Munsky, Albright, Bailey, Tfaily and Dunbar.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Al-Rfou R., Alain G., Almahairi A., Angermüller C., Bahdanau D., Ballas N., et al. (2016). Theano: a Python framework for fast computation of mathematical expressions. ArXiv [Preprint] arXiv:1605.0268,
-
- Bailey V. L., Smith J. L., Bolton H. (2006). 14C Cycling in lignocellulose-amended soils: predicting long-term C fate from short-term indicators. Biol. Fertil. Soils 42 198–206. 10.1007/s00374-005-0016-y - DOI
LinkOut - more resources
Full Text Sources

