Fine-tuning the spike: role of the nature and topology of the glycan shield in the structure and dynamics of the SARS-CoV-2 S
- PMID: 35126971
- PMCID: PMC8729800
- DOI: 10.1039/d1sc04832e
Fine-tuning the spike: role of the nature and topology of the glycan shield in the structure and dynamics of the SARS-CoV-2 S
Abstract
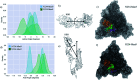
The dense glycan shield is an essential feature of the SARS-CoV-2 spike (S) architecture, key to immune evasion and to the activation of the prefusion conformation. Recent studies indicate that the occupancy and structures of the SARS-CoV-2 S glycans depend not only on the nature of the host cell, but also on the structural stability of the trimer; a point that raises important questions about the relative competence of different glycoforms. Moreover, the functional role of the glycan shield in the SARS-CoV-2 pathogenesis suggests that the evolution of the sites of glycosylation is potentially intertwined with the evolution of the protein sequence to affect optimal activity. Our results from multi-microsecond molecular dynamics simulations indicate that the type of glycosylation at N234, N165 and N343 greatly affects the stability of the receptor binding domain (RBD) open conformation, and thus its exposure and accessibility. Furthermore, our results suggest that the loss of glycosylation at N370, a newly acquired modification in the SARS-CoV-2 S glycan shield's topology, may have contributed to increase the SARS-CoV-2 infectivity as we find that N-glycosylation at N370 stabilizes the closed RBD conformation by binding a specific cleft on the RBD surface. We discuss how the absence of the N370 glycan in the SARS-CoV-2 S frees the RBD glycan binding cleft, which becomes available to bind cell-surface glycans, and potentially increases host cell surface localization.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures



References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

