Discovery of novel selective PI3Kγ inhibitors through combining machine learning-based virtual screening with multiple protein structures and bio-evaluation
- PMID: 35127160
- PMCID: PMC8800018
- DOI: 10.1016/j.jare.2021.04.007
Discovery of novel selective PI3Kγ inhibitors through combining machine learning-based virtual screening with multiple protein structures and bio-evaluation
Abstract
Introduction: Phosphoinositide 3-kinase gamma (PI3Kγ) has been regarded as a promising drug target for the treatment of various diseases, and the diverse physiological roles of class I PI3K isoforms (α, β, δ, and γ) highlight the importance of isoform selectivity in the development of PI3Kγ inhibitors. However, the high structural conservation among the PI3K family makes it a big challenge to develop selective PI3Kγ inhibitors.
Objectives: A novel machine learning-based virtual screening with multiple PI3Kγ protein structures was developed to discover novel PI3Kγ inhibitors.
Methods: A large chemical database was screened using the virtual screening model, the top-ranked compounds were then subjected to a series of bio-evaluations, which led to the discovery of JN-KI3. The selective inhibition mechanism of JN-KI3 against PI3Kγ was uncovered by a theoretical study.
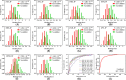
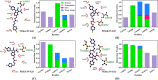
Results: 49 hits were identified through virtual screening, and the cell-free enzymatic studies found that JN-KI3 selectively inhibited PI3Kγ at a concentration as low as 3,873 nM but had no inhibitory effect on Class IA PI3Ks, leading to the selective cytotoxicity on hematologic cancer cells. Meanwhile, JN-KI3 potently blocked the PI3K signaling, finally led to distinct apoptosis of hematologic cell lines at a low concentration. Lastly, the key residues of PI3Kγ and the structural characteristics of JN-KI3, which both would influence γ isoform-selective inhibition, were highlighted by systematic theoretical studies.
Conclusion: The developed virtual screening model strongly manifests the robustness to find novel PI3Kγ inhibitors. JN-KI3 displays a specific cytotoxicity on hematologic tumor cells, and significantly promotes apoptosis associated with the inhibition of the PI3K signaling, which depicts PI3Kγ as a potential target for the hematologic tumor therapy. The theoretical results reveal that those key residues interacting with JN-KI3 are less common compared to most of the reported PI3Kγ inhibitors, indicating that JN-KI3 has novel structural characteristics as a selective PIK3γ inhibitor.
Keywords: ADMET, absorption, distribution, metabolism, excretion, and toxicity; AKT, protein kinase B; AUC, area under receiver operations characteristic curve; Badapple, bioactivity data associative promiscuity pattern learning engine; CADD, computer-aided drug design; CDRA, confirmatory dose–response assays; DMEM, Dulbecco’s Modified Eagle Medium; DS3.5, discovery studio 3.5; FBS, fetal bovine serum; GPCR, G protein-coupled receptors; H-bond, hydrogen bond; Hematologic malignancies; IMDM, Iscove’s Modified Dulbecco’s Medium; Ionic, ionic interactions; JN-KI3; MD, molecular dynamics; MM/GBSA, molecular mechanics/generalized born surface area; Molecular dynamics simulation; NBC, naive Bayesian classifier; PAGE, polyacrylamide gel electrophoresis; PAINS, pan-assay interference compounds; PARP, poly ADP-ribose polymerase; PDB, protein data bank; PI3K, Phosphoinositide 3-kinase; PI3Kγ; PSA, primary screening assays; REOS, rapid elimination of swill; RMSD, root-mean-squared-deviation; RMSF, root-mean-squared-fluctuation; ROC, receiver operations characteristic; RTK, receptor tyrosine kinases; SD, standard deviation; SMILES, simplified molecular input line entry specification; SP, standard precision; Selective inhibitor; VS, virtual screening; Virtual screening; Water Bridge, hydrogen bonds through water molecular bridge; XP, extra precision.
© 2021 The Authors. Published by Elsevier B.V. on behalf of Cairo University.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

