MUC1-C integrates type II interferon and chromatin remodeling pathways in immunosuppression of prostate cancer
- PMID: 35127252
- PMCID: PMC8812775
- DOI: 10.1080/2162402X.2022.2029298
MUC1-C integrates type II interferon and chromatin remodeling pathways in immunosuppression of prostate cancer
Abstract
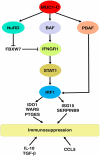
The oncogenic MUC1-C protein drives dedifferentiation of castrate resistant prostate cancer (CRPC) cells in association with chromatin remodeling. The present work demonstrates that MUC1-C is necessary for expression of IFNGR1 and activation of the type II interferon-gamma (IFN-γ) pathway. We show that MUC1-C→ARID1A/BAF signaling induces IFNGR1 transcription and that MUC1-C-induced activation of the NuRD complex suppresses FBXW7 in stabilizing the IFNGR1 protein. MUC1-C and NuRD were also necessary for expression of the downstream STAT1 and IRF1 transcription factors. We further demonstrate that MUC1-C and PBRM1/PBAF are necessary for IRF1-induced expression of (i) IDO1, WARS and PTGES, which metabolically suppress the immune tumor microenvironment (TME), and (ii) the ISG15 and SERPINB9 inhibitors of T cell function. Of translational relevance, we show that MUC1 associates with expression of IFNGR1, STAT1 and IRF1, as well as the downstream IDO1, WARS, PTGES, ISG15 and SERPINB9 immunosuppressive effectors in CRPC tumors. Analyses of scRNA-seq data further demonstrate that MUC1 correlates with cancer stem cell (CSC) and IFN gene signatures across CRPC cells. Consistent with these results, MUC1 associates with immune cell-depleted "cold" CRPC TMEs. These findings demonstrate that MUC1-C integrates chronic activation of the type II IFN-γ pathway and induction of chromatin remodeling complexes in linking the CSC state with immune evasion.
Keywords: CRPC; IFN-gamma; MUC1-C; PBRM1; immunosuppression.
© 2022 The Author(s). Published with license by Taylor & Francis Group, LLC.
Conflict of interest statement
DK has equity interests in Genus Oncology, Reata Pharmaceuticals, and HillstreamBioPharma, and is a paid consultant to Reata and CanBas.
Figures







References
-
- Aggarwal R, Huang J, Alumkal JJ, Zhang L, Feng FY, Thomas GV, Weinstein AS, Friedl V, Zhang C, Witte ON, et al. Clinical and genomic characterization of treatment-emergent small-cell neuroendocrine prostate cancer: a multi-institutional prospective study. J Clin Oncol. 2018;36(24):2492–2503. doi:10.1200/JCO.2017.77.6880. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
