Therapeutic regulation of autophagy in hepatic metabolism
- PMID: 35127371
- PMCID: PMC8799888
- DOI: 10.1016/j.apsb.2021.07.021
Therapeutic regulation of autophagy in hepatic metabolism
Abstract
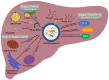
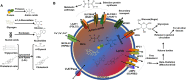
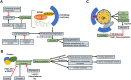
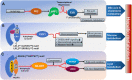
Metabolic homeostasis requires dynamic catabolic and anabolic processes. Autophagy, an intracellular lysosomal degradative pathway, can rewire cellular metabolism linking catabolic to anabolic processes and thus sustain homeostasis. This is especially relevant in the liver, a key metabolic organ that governs body energy metabolism. Autophagy's role in hepatic energy regulation has just begun to emerge and autophagy seems to have a much broader impact than what has been appreciated in the field. Though classically known for selective or bulk degradation of cellular components or energy-dense macromolecules, emerging evidence indicates autophagy selectively regulates various signaling proteins to directly impact the expression levels of metabolic enzymes or their upstream regulators. Hence, we review three specific mechanisms by which autophagy can regulate metabolism: A) nutrient regeneration, B) quality control of organelles, and C) signaling protein regulation. The plasticity of the autophagic function is unraveling a new therapeutic approach. Thus, we will also discuss the potential translation of promising preclinical data on autophagy modulation into therapeutic strategies that can be used in the clinic to treat common metabolic disorders.
Keywords: AIM, Atf8 interacting motif; ATGL, adipose triglyceride lipase; ATL3, Atlastin GTPase 3; ATM, ATM serine/threonine kinase; Autophagy; BA, bile acid; BCL2L13, BCL2 like 13; BNIP3, BCL2 interacting protein 3; BNIP3L, BCL2 interacting protein 3 like; CAR, constitutive androstane receptor; CCPG1, cell cycle progression 1; CLN3, lysosomal/endosomal transmembrane protein; CMA, chaperonin mediated autophagy; CREB, cAMP response element binding protein; CRY1, cryptochrome 1; CYP27A1, sterol 27-hydroxylase; CYP7A1, cholesterol 7α-hydroxylase; Cryptochrome 1; DFCP1, double FYVE-containing protein 1; FAM134B, family with sequence similarity 134, member B; FFA, free fatty acid; FOXO1, Forkhead box O1; FUNDC1, FUN14 domain containing 1; FXR, farnesoid X receptor; Farnesoid X receptor; GABARAPL1, GABA type A receptor associated protein like 1; GIM, GABARAP-interacting motif; LAAT-1, lysosomal amino acid transporter 1 homologue; LALP70, lysosomal apyrase-like protein of 70 kDa; LAMP1, lysosomal-associated membrane protein-1; LAMP2, lysosomal-associated membrane protein-2; LD, lipid droplet; LIMP1, lysosomal integral membrane protein-1; LIMP3, lysosomal integral membrane protein-3; LIR, LC3 interacting region; LXRa, liver X receptor a; LYAAT-1, lysosomal amino acid transporter 1; Liver metabolism; Lysosome; MCOLN1, mucolipin 1; MFSD1, major facilitator superfamily domain containing 1; NAFLD, non-alcoholic fatty liver disease; NBR1, BRCA1 gene 1 protein; NCoR1, nuclear receptor co-repressor 1; NDP52, calcium-binding and coiled-coil domain-containing protein 2; NPC-1, Niemann-Pick disease, type C1; Nutrient regeneration; OPTN, optineurin; PEX5, peroxisomal biogenesis factor 5; PI3K, phosphatidylinositol-4,5-bisphosphate 3-kinase; PINK1, phosphatase and tensin homolog (PTEN)-induced kinase 1; PKA, protein kinase A; PKB, protein kinase B; PLIN2, perilipin 2; PLIN3, perilipin 3; PP2A, protein phosphatase 2a; PPARα, peroxisomal proliferator-activated receptor-alpha; PQLC2, PQ-loop protein; PXR, pregnane X receptor; Quality control; RETREG1, reticulophagy regulator 1; ROS, reactive oxygen species; RTN3, reticulon 3; RTNL3, a long isoform of RTN3; S1PR2, sphingosine-1-phosphate receptor 2; S6K, P70-S6 kinase; S6RP, S6 ribosomal protein; SCARB2, scavenger receptor class B member 2; SEC62, SEC62 homolog, preprotein translocation factor; SIRT1, sirtuin 1; SLC36A1, solute carrier family 36 member 1; SLC38A7, solute carrier family 38 member 7; SLC38A9, sodium-coupled neutral amino acid transporter 9; SNAT7, sodium-coupled neutral amino acid transporter 7; SPIN, spindling; SQSTM1, sequestosome 1; STBD1, starch-binding domain-containing protein 1; Signaling proteins; TBK1, serine/threonine-protein kinase; TEX264, testis expressed 264, ER-phagy receptor; TFEB/TFE3, transcription factor EB; TGR5, takeda G protein receptor 5; TRAC-1, thyroid-hormone-and retinoic acid-receptor associated co-repressor 1; TRPML1, transient receptor potential mucolipin 1; ULK1, Unc-51 like autophagy activating kinase 1; UPR, unfolded protein response; V-ATPase, vacuolar-ATPase; VDR, vitamin D3 receptor; VLDL, very-low-density lipoprotein; WIPI1, WD repeat domain phosphoinositide-interacting protein 1; mTORC1, mammalian target of rapamycin complex 1.
© 2022 Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences. Production and hosting by Elsevier B.V.
Figures

References
-
- Zhao J., Benlekbir S., Rubinstein J.L. Electron cryomicroscopy observation of rotational states in a eukaryotic V-ATPase. Nature. 2015;521:241–245. - PubMed
-
- Mellman I. Organelles observed: lysosomes. Science. 1989;244:853–854. - PubMed
-
- Neiss W.F. A coat of glycoconjugates on the inner surface of the lysosomal membrane in the rat kidney. Histochemistry. 1984;80:603–608. - PubMed
-
- Eskelinen E.L., Tanaka Y., Saftig P. At the acidic edge: emerging functions for lysosomal membrane proteins. Trends Cell Biol. 2003;13:137–145. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

