Anti-Tumor Role of CAMK2B in Remodeling the Stromal Microenvironment and Inhibiting Proliferation in Papillary Renal Cell Carcinoma
- PMID: 35127542
- PMCID: PMC8815460
- DOI: 10.3389/fonc.2022.740051
Anti-Tumor Role of CAMK2B in Remodeling the Stromal Microenvironment and Inhibiting Proliferation in Papillary Renal Cell Carcinoma
Abstract
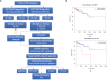
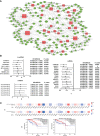
The tumor microenvironment (TME) is variable across tumor types and has diverse effects on malignant progression, based on the type and number of infiltrating stromal cells. In particular, TME effector genes and their competitive endogenous RNA (ceRNA) networks play a critical role in regulating malignant tumor progression. However, the core effector molecules involved in TME modulation of kidney renal papillary cell carcinoma (KIRP) are poorly understood. To address this question, a cohort containing 233 KIRP patients was derived from The Cancer Genome Atlas (TCGA) database, and the data were processed using the ESTIMATE algorithm. We further evaluated the relationship between immune scores (ISs) and stromal scores (SSs) and disease progression and found that high SSs were associated with a poor prognosis in KIRP. Differentially expressed genes (DEGs) were therefore screened based on SS scores, resulting in 2509 DEGs, including 1668 mRNAs, 783 long noncoding (lnc)RNAs, and 58 micro (mi)RNAs. DEGs were then filtered using the random variance and subjected to hierarchical clustering using EPCLUST. Weighted gene co-expression network analysis (WGCNA) was used to assess the prognostic capacity of these DEGs and identify target ceRNA networks, and lncRNA GUSBP11/miR-432-5p/CAMK2B in the turquoise module was selected as a promising ceRNA network. From this analysis CAMK2B was selected as the core gene predicted to be involved in stromal TMA regulation. We therefore explored the expression and function of CAMK2B in vitro and in vivo and provide evidence that this protein promotes stromal TME remodulation and inhibits proliferation in KIRP. Lastly, we show that vascular endothelial growth factor (VEGF), transforming growth factor (TGF)β, and close homolog of L1 (CHL1) act as downstream effectors of CAMK2B in KIRP. Thus, in this study, we show that the TME determines prognosis of KIRP patients via the core effector molecule CAMK2B, which mediates both microenvironmental remodeling and tumor progression. Based on these findings, we propose that remodeling of the stromal microenvironment could represent an improved therapeutic approach relative to immunotherapy for KIRP.
Keywords: CAMK2B; kidney renal papillary cell carcinoma (KIRP); lncRNA GUSBP11; miR-432-5p; tumor microenvironment.
Copyright © 2022 Jia, Liao, Zhang, Xu, Song, Bian and Fu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
LinkOut - more resources
Full Text Sources
Miscellaneous

