Identification and Validation of a Novel Genomic Instability-Associated Long Non-Coding RNA Prognostic Signature in Head and Neck Squamous Cell Carcinoma
- PMID: 35127708
- PMCID: PMC8812830
- DOI: 10.3389/fcell.2021.787766
Identification and Validation of a Novel Genomic Instability-Associated Long Non-Coding RNA Prognostic Signature in Head and Neck Squamous Cell Carcinoma
Abstract
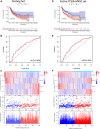
Background: Head and neck squamous cell carcinoma (HNSCC) is one of the most aggressive malignant cancers worldwide, and accurate prognostic models are urgently needed. Emerging evidence revealed that long non-coding RNAs (lncRNAs) are related to genomic instability. We sought to identify and validate a genomic instability-associated lncRNA prognostic signature to assess HNSCC patient survival outcomes. Methods: RNA-sequencing data, somatic mutation files, and patient clinical data were downloaded from The Cancer Genome Atlas database. A total of 491 patients with completely clinical files were randomly divided into training and testing sets. In the training set, genomic instability-associated lncRNAs were screened through univariate Cox regression analyses and least absolute shrinkage and selection operator regression analyses to build a genomic instability-associated lncRNA signature (GILncSig). In addition, time-dependent receiver operating characteristic (ROC) curve, Kaplan-Meier survival curve, and clinical stratification analyses were used to evaluate the signature's reliability. Finally, in situ hybridization experiments were performed to validate GILncSig expression levels between adjacent non-tumor tissues and tumor tissues from HNSCC patients. Results: Four genomic instability-associated lncRNAs (AC023310.4, AC091729.1, LINC01564, and MIR3142HG) were selected for the prognostic signature. The model was successfully validated using the testing cohort. ROC analysis demonstrated its strong predictive ability for HNSCC prognosis. Univariate and multivariate Cox analyses revealed that the GILncSig was an independent predictor of prognosis. HNSCC patients with a low-risk score showed a substantially better prognosis than the high-risk groups. The in situ hybridization experiments using human HNSCC tissue revealed high GILncSig expression in HNSCC tissues compared with adjacent non-tumor tissues. Conclusion: We developed a novel GILncSig for prognosis prediction in HNSCC patients, and the components of that signature might be therapeutic targets for HNSCC.
Keywords: genomic instability; head and neck squamous cell carcinoma; long non-coding RNA; prognostic signature; survival.
Copyright © 2022 Chen, Zhao, Lu, Zhao and Guo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







References
-
- Bao S., Zhao H., Yuan J., Fan D., Zhang Z., Su J., et al. (2020). Computational Identification of Mutator-Derived lncRNA Signatures of Genome Instability for Improving the Clinical Outcome of Cancers: a Case Study in Breast Cancer. Brief. Bioinformatics 21, 1742–1755. 10.1093/bib/bbz118 - DOI - PubMed
LinkOut - more resources
Full Text Sources

