Identification of Potential Key Genes and Molecular Mechanisms of Medulloblastoma Based on Integrated Bioinformatics Approach
- PMID: 35127939
- PMCID: PMC8816556
- DOI: 10.1155/2022/1776082
Identification of Potential Key Genes and Molecular Mechanisms of Medulloblastoma Based on Integrated Bioinformatics Approach
Retraction in
-
Retracted: Identification of Potential Key Genes and Molecular Mechanisms of Medulloblastoma Based on Integrated Bioinformatics Approach.Biomed Res Int. 2024 Mar 20;2024:9853643. doi: 10.1155/2024/9853643. eCollection 2024. Biomed Res Int. 2024. PMID: 38550031 Free PMC article.
Abstract
Background: Medulloblastoma (MB) is the most occurring brain cancer that mostly happens in childhood age. This cancer starts in the cerebellum part of the brain. This study is designed to screen novel and significant biomarkers, which may perform as potential prognostic biomarkers and therapeutic targets in MB.
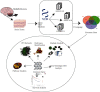
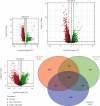
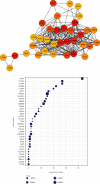
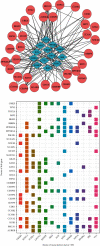
Methods: A total of 103 MB-related samples from three gene expression profiles of GSE22139, GSE37418, and GSE86574 were downloaded from the Gene Expression Omnibus (GEO). Applying the limma package, all three datasets were analyzed, and 1065 mutual DEGs were identified including 408 overexpressed and 657 underexpressed with the minimum cut-off criteria of ∣log fold change | >1 and P < 0.05. The Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and WikiPathways enrichment analyses were executed to discover the internal functions of the mutual DEGs. The outcomes of enrichment analysis showed that the common DEGs were significantly connected with MB progression and development. The Search Tool for Retrieval of Interacting Genes (STRING) database was used to construct the interaction network, and the network was displayed using the Cytoscape tool and applying connectivity and stress value methods of cytoHubba plugin 35 hub genes were identified from the whole network.
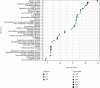
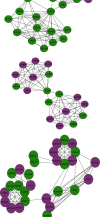
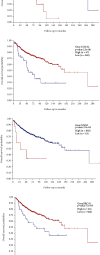
Results: Four key clusters were identified using the PEWCC 1.0 method. Additionally, the survival analysis of hub genes was brought out based on clinical information of 612 MB patients. This bioinformatics analysis may help to define the pathogenesis and originate new treatments for MB.
Copyright © 2022 Md. Rakibul Islam et al.
Conflict of interest statement
All the authors have read the manuscript and approved this for submission; also, there are no competing interests.
Figures



References
-
- Kool M., Korshunov A., Remke M., et al. Molecular subgroups of medulloblastoma: an international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, group 3, and group 4 medulloblastomas. Actaneuropathologica . 2012;123(4):473–484. doi: 10.1007/s00401-012-0958-8. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

