BepiTBR: T-B reciprocity enhances B cell epitope prediction
- PMID: 35128358
- PMCID: PMC8803616
- DOI: 10.1016/j.isci.2022.103764
BepiTBR: T-B reciprocity enhances B cell epitope prediction
Abstract
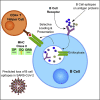
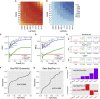
The ability to predict B cell epitopes is critical for biomedical research and many clinical applications. Investigators have observed the phenomenon of T-B reciprocity, in which candidate B cell epitopes with nearby CD4+ T cell epitopes have higher chances of being immunogenic. To our knowledge, existing B cell epitope prediction algorithms have not considered this interesting observation. We developed a linear B cell epitope prediction model, BepiTBR, based on T-B reciprocity. We showed that explicitly including the enrichment of putative CD4+ T cell epitopes (predicted HLA class II epitopes) in the model leads to significant enhancement in the prediction of linear B cell epitopes. Curiously, the positive impact on B cell epitope generation is specific to the enrichment of DQ allele binders. Overall, our work provides interesting mechanistic insights into the generation of B cell epitopes and points to a new avenue to improve B cell epitope prediction for the field.
Keywords: Bioinformatics; Immunology; Systems biology.
© 2022.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Benjamin D.C., Berzofsky J.A., East I.J., Gurd F.R., Hannum C., Leach S.J., Margoliash E., Michael J.G., Miller A., Prager E.M. The antigenic structure of proteins: a reappraisal. Annu. Rev. Immunol. 1984;2:67–101. - PubMed
-
- Berzofsky J.A. T-B reciprocity. An Ia-restricted epitope-specific circuit regulating T cell-B cell interaction and antibody specificity. Surv. Immunol. Res. 1983;2:223–229. - PubMed
-
- Berzofsky J.A., Buckenmeyer G.K., Hicks G., Gurd F.R., Feldmann R.J., Minna J. Topographic antigenic determinants recognized by monoclonal antibodies to sperm whale myoglobin. J. Biol. Chem. 1982;257:3189–3198. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

