Clinical validation of genomic functional screen data: Analysis of observed BRCA1 variants in an unselected population cohort
- PMID: 35128484
- PMCID: PMC8804171
- DOI: 10.1016/j.xhgg.2022.100086
Clinical validation of genomic functional screen data: Analysis of observed BRCA1 variants in an unselected population cohort
Abstract
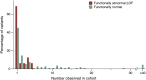
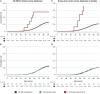
Functional assessment of genomic variants provides a promising approach to systematically examine the potential pathogenicity of variants independent of associated clinical data. However, making such conclusions requires validation with appropriate clinical findings. To this end, here, we use variant calls from exome data and BRCA1-related cancer diagnoses from electronic health records to demonstrate an association between published laboratory-based functional designations of BRCA1 variants and BRCA1-related cancer diagnoses in an unselected cohort of patient-participants. These findings validate and support further exploration of functional assay data to better understand the pathogenicity of rare variants. This information may be valuable in the context of healthy population genomic screening, where many rare, potentially pathogenic variants may not have sufficient associated clinical data to inform their interpretation directly.
Keywords: BRCA1; electronic health record; functional screen; hereditary breast and ovarian cancer; population genomics; rare variant interpretation.
© 2022 The Author(s).
Conflict of interest statement
K.M.S.B. is an employee at Helix. M.M.’s affiliation with The MITRE Corporation is provided for identification purposes only and is not intended to convey or imply MITRE’s concurrence with, or support for, the positions, opinions, or viewpoints expressed by M.M. K.E.H. is an employee of and shareholder in Invitae. H.F.W. is an employee at Genome Medical.
Figures
References
-
- Strande N.T., Riggs E.R., Buchanan A.H., Ceyhan-Birsoy O., DiStefano M., Dwight S.S., Goldstein J., Ghosh R., Seifert B.A., Sneddon T.P., et al. Evaluating the clinical validity of gene-disease associations: an evidence-based framework developed by the Clinical Genome Resource. Am. J. Hum. Genet. 2017;100:895–906. - PMC - PubMed
-
- Stenson P.D., Mort M., Ball E.V., Evans K., Hayden M., Heywood S., Hussain M., Phillips A.D., Cooper D.N. The human gene mutation database: towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum. Genet. 2017;136:665–677. - PMC - PubMed
-
- Rehm H.L., Berg J.S., Plon S.E. ClinGen and ClinVar - enabling genomics in precision medicine. Hum. Mutat. 2018;39:1473–1475.
LinkOut - more resources
Full Text Sources
Miscellaneous

