Leveraging TOPMed imputation server and constructing a cohort-specific imputation reference panel to enhance genotype imputation among cystic fibrosis patients
- PMID: 35128485
- PMCID: PMC8804187
- DOI: 10.1016/j.xhgg.2022.100090
Leveraging TOPMed imputation server and constructing a cohort-specific imputation reference panel to enhance genotype imputation among cystic fibrosis patients
Abstract
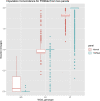
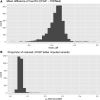
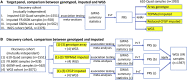
Cystic fibrosis (CF) is a severe genetic disorder that can cause multiple comorbidities affecting the lungs, the pancreas, the luminal digestive system and beyond. In our previous genome-wide association studies (GWAS), we genotyped approximately 8,000 CF samples using a mixture of different genotyping platforms. More recently, the Cystic Fibrosis Genome Project (CFGP) performed deep (approximately 30×) whole genome sequencing (WGS) of 5,095 samples to better understand the genetic mechanisms underlying clinical heterogeneity among patients with CF. For mixtures of GWAS array and WGS data, genotype imputation has proven effective in increasing effective sample size. Therefore, we first performed imputation for the approximately 8,000 CF samples with GWAS array genotype using the Trans-Omics for Precision Medicine (TOPMed) freeze 8 reference panel. Our results demonstrate that TOPMed can provide high-quality imputation for patients with CF, boosting genomic coverage from approximately 0.3-4.2 million genotyped markers to approximately 11-43 million well-imputed markers, and significantly improving polygenic risk score (PRS) prediction accuracy. Furthermore, we built a CF-specific CFGP reference panel based on WGS data of patients with CF. We demonstrate that despite having approximately 3% the sample size of TOPMed, our CFGP reference panel can still outperform TOPMed when imputing some CF disease-causing variants, likely owing to allele and haplotype differences between patients with CF and general populations. We anticipate our imputed data for 4,656 samples without WGS data will benefit our subsequent genetic association studies, and the CFGP reference panel built from CF WGS samples will benefit other investigators studying CF.
Keywords: cystic fibrosis; genotype imputation; mendelian disease; polygenic risk score.
© 2022 The Authors.
Conflict of interest statement
M.J.B. is the Editor-in-chief of HGG Advances. All other authors declare no competing interests.
Figures

References
-
- Aksit M.A., Pace R.G., Vecchio-Pagán B., Ling H., Rommens J.M., Boelle P.-Y., Guillot L., Raraigh K.S., Pugh E., Zhang P., et al. Genetic modifiers of cystic fibrosis-related diabetes have extensive overlap with type 2 diabetes and related traits. J. Clin. Endocrinol. Metab. 2020;105:1401–1415. - PMC - PubMed
-
- Treggiari M.M., Rosenfeld M., Mayer-Hamblett N., Retsch-Bogart G., Gibson R.L., Williams J., Emerson J., Kronmal R.A., Ramsey B.W. Early anti-pseudomonal acquisition in young patients with cystic fibrosis: rationale and design of the EPIC clinical trial and observational study. Contemp. Clin. Trials. 2009;30:256–268. - PMC - PubMed
-
- Kowalski M.H., Qian H., Hou Z., Rosen J.D., Tapia A.L., Shan Y., Jain D., Argos M., Arnett D.K., Avery C., et al. Use of >100,000 NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium whole genome sequences improves imputation quality and detection of rare variant associations in admixed African and Hispanic/Latino populations. PLoS Genet. 2019;15:e1008500. - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

