Selenium-Dependent Read Through of the Conserved 3'-Terminal UGA Stop Codon of HIV-1 nef
- PMID: 35128545
- PMCID: PMC8813066
- DOI: 10.25259/ajbps_6_2021
Selenium-Dependent Read Through of the Conserved 3'-Terminal UGA Stop Codon of HIV-1 nef
Abstract
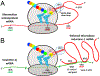
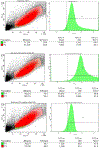
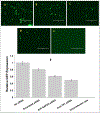
The HIV-1 nef gene terminates in a 3'-UGA stop codon, which is highly conserved in the main group of HIV-1 subtypes, along with a downstream potential coding region that could extend the nef protein by 33 amino acids, if readthrough of the stop codon occurs. Antisense tethering interactions (ATIs) between a viral mRNA and a host selenoprotein mRNA are a potential viral strategy for the capture of a host selenocysteine insertion sequence (SECIS) element (Taylor et al, 2016) [1]. This mRNA hijacking mechanism could enable the expression of virally encoded selenoprotein modules, via translation of in-frame UGA stop codons as selenocysteine (SeC). Here we show that readthrough of the 3'-terminal UGA codon of nef occurs during translation of HIV-1 nef expression constructs in transfected cells. This was accomplished via fluorescence microscopy image analysis and flow cytometry of HEK 293 cells, transfected with engineered GFP reporter gene plasmid constructs, in which GFP can only be expressed by translational recoding of the UGA codon. SiRNA knockdown of thioredoxin reductase 1 (TR1) mRNA resulted in a 67% decrease in GFP expression, presumably due to reduced availability of the components involved in selenocysteine incorporation for the stop codon readthrough, thus supporting the proposed ATI. Addition of 20 nM sodium selenite to the media significantly enhanced stop codon readthrough in the pNefATI1 plasmid construct, by >100%, supporting the hypothesis that selenium is involved in the UGA readthrough mechanism.
Conflict of interest statement
Conflicts of Interest All authors declare that they have no conflicts of interest.
Figures
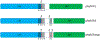




References
-
- Zhong L; Arner ES; Holmgren A Structure and mechanism of mammalian thioredoxin reductase: the active site is a redox-active selenolthiol/selenenylsulfide formed from the conserved cysteine-selenocysteine sequence. Proc Natl Acad Sci U S A 2000, 97, 5854–5859, doi: 10.1073/pnas.100114897. - DOI - PMC - PubMed
-
- Taylor EW; Cox AG; Zhao L; Ruzicka JA; Bhat AA; Zhang W; Nadimpalli RG; Dean RG Nutrition, HIV, and drug abuse: the molecular basis of a unique role for selenium. J Acquir Immune Defic Syndr 2000, 25 Suppl 1, S53–61. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
