Molecular profile of FLT3-mutated relapsed/refractory patients with AML in the phase 3 ADMIRAL study of gilteritinib
- PMID: 35130342
- PMCID: PMC9006281
- DOI: 10.1182/bloodadvances.2021006489
Molecular profile of FLT3-mutated relapsed/refractory patients with AML in the phase 3 ADMIRAL study of gilteritinib
Abstract
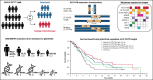
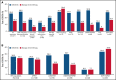
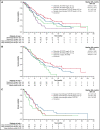
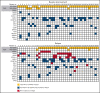
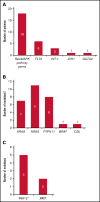
The phase 3 Study of ASP2215 Versus Salvage Chemotherapy in Patients With Relapsed or Refractory Acute Myeloid Leukemia (AML) With FMS-like Tyrosine Kinase (FLT3) Mutation (ADMIRAL) trial demonstrated the superiority of the FLT3 inhibitor, gilteritinib, to salvage chemotherapy (SC) in patients with FLT3-mutated relapsed or refractory (R/R) AML. Baseline comutations, FLT3-internal tandem duplication (ITD) allelic ratio and length, and treatment-emergent mutations were analyzed in patients in the ADMIRAL trial. Baseline comutations were grouped according to gene subgroups (DNA methylation/hydroxymethylation, transcription, chromatin-spliceosome, receptor tyrosine kinase-Ras signaling, TP53-aneuploidy, NPM1, DNMT3A, DNMT3A/NPM1, WT-1, and IDH1/IDH2). Across all but 1 gene subgroup (TP53-aneuploidy), higher pretransplant response rates and a trend toward longer overall survival were observed with gilteritinib vs SC. Patients with DNMT3A/NPM1 comutations who received gilteritinib had the most favorable outcomes of any molecular subgroup analyzed. Survival outcomes with gilteritinib were not adversely affected by FLT3-ITD allelic ratio, FLT3-ITD length, or multiple FLT3-ITD mutations. Among patients who relapsed on gilteritinib, Ras/mitogen-activated protein kinase (MAPK) pathway and FLT3 F691L gene mutations were the most common mutational events associated with treatment resistance. However, the occurrence of Ras/MAPK pathway gene mutations at baseline did not preclude a clinical benefit from gilteritinib. Acquisition of multiple Ras/MAPK pathway gene mutations at relapse suggests a high level of pathway reactivation is needed to overcome the gilteritinib treatment effect. These findings provide insight into the R/R AML molecular profile and the impact of FLT3 inhibitors on mutational evolution associated with treatment resistance and benefit of gilteritinib across a wide spectrum of molecular and genetic subgroups in FLT3-mutated R/R AML. This trial was registered at www.clinicaltrials.gov as #NCT02421939.
© 2022 by The American Society of Hematology. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Figures

References
-
- Thiede C, Steudel C, Mohr B, et al. . Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: association with FAB subtypes and identification of subgroups with poor prognosis. Blood. 2002;99(12):4326-4335. - PubMed
-
- Kottaridis PD, Gale RE, Frew ME, et al. . The presence of a FLT3 internal tandem duplication in patients with acute myeloid leukemia (AML) adds important prognostic information to cytogenetic risk group and response to the first cycle of chemotherapy: analysis of 854 patients from the United Kingdom Medical Research Council AML 10 and 12 trials. Blood. 2001;98(6):1752-1759. - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

