Amplification Artifact in SARS-CoV-2 Omicron Sequences Carrying P681R Mutation, New York, USA
- PMID: 35130474
- PMCID: PMC8962901
- DOI: 10.3201/eid2804.220146
Amplification Artifact in SARS-CoV-2 Omicron Sequences Carrying P681R Mutation, New York, USA
Abstract
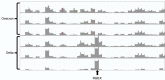
Of 379 severe acute respiratory syndrome coronavirus 2 samples collected in New York, USA, we detected 86 Omicron variant sequences containing Delta variant mutation P681R. Probable explanations were co-infection with 2 viruses or contamination/amplification artifact. Repeated library preparation with fewer cycles showed the P681R calls were artifactual. Unusual mutations should be interpreted with caution.
Keywords: 2019 novel coronavirus disease; COVID-19; SARS-CoV-2; amplification artifact; coronavirus disease; genomics; respiratory infections; sequence analysis; severe acute respiratory syndrome coronavirus 2; viruses; zoonoses.
Figures
References
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

