Pyrimidine inhibitors synergize with nucleoside analogues to block SARS-CoV-2
- PMID: 35130559
- PMCID: PMC10377386
- DOI: 10.1038/s41586-022-04482-x
Pyrimidine inhibitors synergize with nucleoside analogues to block SARS-CoV-2
Abstract
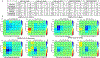
The SARS-CoV-2 virus has infected more than 261 million people and has led to more than 5 million deaths in the past year and a half1 ( https://www.who.org/ ). Individuals with SARS-CoV-2 infection typically develop mild-to-severe flu-like symptoms, whereas infection of a subset of individuals leads to severe-to-fatal clinical outcomes2. Although vaccines have been rapidly developed to combat SARS-CoV-2, there has been a dearth of antiviral therapeutics. There is an urgent need for therapeutics, which has been amplified by the emerging threats of variants that may evade vaccines. Large-scale efforts are underway to identify antiviral drugs. Here we screened approximately 18,000 drugs for antiviral activity using live virus infection in human respiratory cells and validated 122 drugs with antiviral activity and selectivity against SARS-CoV-2. Among these candidates are 16 nucleoside analogues, the largest category of clinically used antivirals. This included the antivirals remdesivir and molnupiravir, which have been approved for use in COVID-19. RNA viruses rely on a high supply of nucleoside triphosphates from the host to efficiently replicate, and we identified a panel of host nucleoside biosynthesis inhibitors as antiviral. Moreover, we found that combining pyrimidine biosynthesis inhibitors with antiviral nucleoside analogues synergistically inhibits SARS-CoV-2 infection in vitro and in vivo against emerging strains of SARS-CoV-2, suggesting a clinical path forward.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Figures

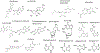
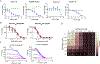






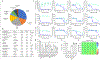



Update of
-
Pyrimidine biosynthesis inhibitors synergize with nucleoside analogs to block SARS-CoV-2 infection.bioRxiv [Preprint]. 2021 Jun 24:2021.06.24.449811. doi: 10.1101/2021.06.24.449811. bioRxiv. 2021. Update in: Nature. 2022 Apr;604(7904):134-140. doi: 10.1038/s41586-022-04482-x. PMID: 34189531 Free PMC article. Updated. Preprint.
Comment in
-
A promising strategy against SARS-CoV-2: pyrimidine inhibitors synergize with nucleoside analogues.Signal Transduct Target Ther. 2022 Mar 15;7(1):88. doi: 10.1038/s41392-022-00956-6. Signal Transduct Target Ther. 2022. PMID: 35292618 Free PMC article. No abstract available.
References
-
- Jordheim LP, Durantel D, Zoulim F.& Dumontet C.Advances in the development of nucleoside and nucleotide analogues for cancer and viral diseases. Nat. Rev. Drug Discov 12, 447–464 (2013). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

