Subcellular Detection of SARS-CoV-2 RNA in Human Tissue Reveals Distinct Localization in Alveolar Type 2 Pneumocytes and Alveolar Macrophages
- PMID: 35130722
- PMCID: PMC8822351
- DOI: 10.1128/mbio.03751-21
Subcellular Detection of SARS-CoV-2 RNA in Human Tissue Reveals Distinct Localization in Alveolar Type 2 Pneumocytes and Alveolar Macrophages
Abstract
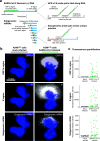
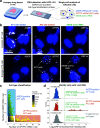
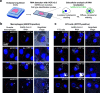
The widespread coronavirus disease 2019 (COVID-19) is caused by infection with the novel coronavirus SARS-CoV-2. Currently, we have limited understanding of which cells become infected with SARS-CoV-2 in human tissues and where viral RNA localizes on the subcellular level. Here, we present a platform for preparing autopsy tissue for visualizing SARS-CoV-2 RNA using RNA fluorescence in situ hybridization (FISH) with amplification by hybridization chain reaction. We developed probe sets that target different regions of SARS-CoV-2 (including ORF1a and N), as well as probe sets that specifically target SARS-CoV-2 subgenomic mRNAs. We validated these probe sets in cell culture and tissues (lung, lymph node, and placenta) from infected patients. Using this technology, we observe distinct subcellular localization patterns of the ORF1a and N regions. In human lung tissue, we performed multiplexed RNA FISH HCR for SARS-CoV-2 and cell-type-specific marker genes. We found viral RNA in cells containing the alveolar type 2 (AT2) cell marker gene (SFTPC) and the alveolar macrophage marker gene (MARCO) but did not identify viral RNA in cells containing the alveolar type 1 (AT1) cell marker gene (AGER). Moreover, we observed distinct subcellular localization patterns of viral RNA in AT2 cells and alveolar macrophages. In sum, we demonstrate the use of RNA FISH HCR for visualizing different RNA species from SARS-CoV-2 in cell lines and FFPE (formalin fixation and paraffin embedding) autopsy specimens. We anticipate that this platform could be broadly useful for studying SARS-CoV-2 pathology in tissues, as well as extended for other applications, including investigating the viral life cycle, viral diagnostics, and drug screening. IMPORTANCE Here, we developed an in situ RNA detection assay for RNA generated by the SARS-CoV-2 virus. We found viral RNA in lung, lymph node, and placenta samples from pathology specimens from COVID patients. Using high-magnification microscopy, we can visualize the subcellular distribution of these RNA in single cells.
Keywords: RNA FISH; cellular imaging; fluorescent image analysis; single cell.
Conflict of interest statement
The authors declare a conflict of interest. S.M.S. receives royalties related to Stellaris RNA FISH probes. All other authors declare no competing interests.
S.M.S. receives royalties related to Stellaris RNA FISH probes. All other authors declare no competing interests.
Figures

References
-
- Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. 2020. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395:497–506. doi:10.1016/S0140-6736(20)30183-5. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
