Novel CRK-Cyclin Complex Controls Spindle Assembly Checkpoint in Toxoplasma Endodyogeny
- PMID: 35130726
- PMCID: PMC8822342
- DOI: 10.1128/mbio.03561-21
Novel CRK-Cyclin Complex Controls Spindle Assembly Checkpoint in Toxoplasma Endodyogeny
Abstract
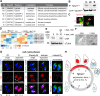
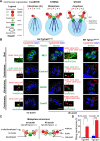
Opportunistic parasites of the Apicomplexa phylum use a variety of division modes built on two types of cell cycles that incorporate two distinctive mechanisms of mitosis: uncoupled from and coupled to parasite budding. Parasites have evolved novel factors to regulate such unique replication mechanisms that are poorly understood. Here, we have combined genetics, quantitative fluorescence microscopy, and global proteomics approaches to examine endodyogeny in Toxoplasma gondii dividing by mitosis coupled to cytokinesis. In the current study, we focus on the steps controlled by the recently described atypical Cdk-related kinase T. gondii Crk6 (TgCrk6). While inspecting protein complexes, we found that this previously orphaned TgCrk6 kinase interacts with a parasite-specific atypical cyclin, TgCyc1. We built conditional expression models and examined primary cell cycle defects caused by the lack of TgCrk6 or TgCyc1. Quantitative microscopy assays revealed that tachyzoites deficient in either TgCrk6 or the cyclin partner TgCyc1 exhibit identical mitotic defects, suggesting cooperative action of the complex components. Further examination of the mitotic structures indicated that the TgCrk6/TgCyc1 complex regulates metaphase. This novel finding confirms a functional spindle assembly checkpoint (SAC) in T. gondii. Measuring global changes in protein expression and phosphorylation, we found evidence that canonical activities of the Toxoplasma SAC are intertwined with parasite-specific tasks. Analysis of phosphorylation motifs suggests that Toxoplasma metaphase is regulated by CDK, mitogen-activated kinase (MAPK), and Aurora kinases, while the TgCrk6/TgCyc1 complex specifically controls the centromere-associated network. IMPORTANCE The rate of Toxoplasma tachyzoite division directly correlates with the severity of the disease, toxoplasmosis, which affects humans and animals. Thus, a better understanding of the tachyzoite cell cycle would offer much-needed efficient tools to control the acute stage of infection. Although tachyzoites divide by binary division, the cell cycle architecture and regulation differ significantly from the conventional binary fission of their host cells. Unlike the unidirectional conventional cell cycle, the Toxoplasma budding cycle is braided and is regulated by multiple essential Cdk-related kinases (Crks) that emerged in the place of missing conventional cell cycle regulators. How these novel Crks control apicomplexan cell cycles is largely unknown. Here, we have discovered a novel parasite-specific complex, TgCrk6/TgCyc1, that orchestrates a major mitotic event, the spindle assembly checkpoint. We demonstrated that tachyzoites incorporated parasite-specific tasks in the canonical checkpoint functions.
Keywords: Apicomplexa; Toxoplasma gondii; apicomplexan parasites; cell cycle; cyclin; cyclin-dependent kinase; endodyogeny; mitosis; protein phosphorylation; spindle assembly checkpoint.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Gubbels M-J, Keroack CD, Dangoudoubiyam S, Worliczek HL, Paul AS, Bauwens C, Elsworth B, Engelberg K, Howe DK, Coppens I, Duraisingh MT. 2020. Fussing about fission: defining variety among mainstream and exotic apicomplexan cell division modes. Front Cell Infect Microbiol 10:269. doi:10.3389/fcimb.2020.00269. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

