The applications of DNA methylation as a biomarker in kidney transplantation: a systematic review
- PMID: 35130936
- PMCID: PMC8822833
- DOI: 10.1186/s13148-022-01241-7
The applications of DNA methylation as a biomarker in kidney transplantation: a systematic review
Abstract
Background: Although kidney transplantation improves patient survival and quality of life, long-term results are hampered by both immune- and non-immune-mediated complications. Current biomarkers of post-transplant complications, such as allograft rejection, chronic renal allograft dysfunction, and cutaneous squamous cell carcinoma, have a suboptimal predictive value. DNA methylation is an epigenetic modification that directly affects gene expression and plays an important role in processes such as ischemia/reperfusion injury, fibrosis, and alloreactive immune response. Novel techniques can quickly assess the DNA methylation status of multiple loci in different cell types, allowing a deep and interesting study of cells' activity and function. Therefore, DNA methylation has the potential to become an important biomarker for prediction and monitoring in kidney transplantation.
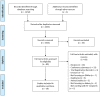
Purpose of the study: The aim of this study was to evaluate the role of DNA methylation as a potential biomarker of graft survival and complications development in kidney transplantation. MATERIAL AND METHODS: A systematic review of several databases has been conducted. The Newcastle-Ottawa scale and the Jadad scale have been used to assess the risk of bias for observational and randomized studies, respectively.
Results: Twenty articles reporting on DNA methylation as a biomarker for kidney transplantation were included, all using DNA methylation for prediction and monitoring. DNA methylation pattern alterations in cells isolated from different tissues, such as kidney biopsies, urine, and blood, have been associated with ischemia-reperfusion injury and chronic renal allograft dysfunction. These alterations occurred in different and specific loci. DNA methylation status has also proved to be important for immune response modulation, having a crucial role in regulatory T cell definition and activity. Research also focused on a better understanding of the role of this epigenetic modification assessment for regulatory T cells isolation and expansion for future tolerance induction-oriented therapies.
Conclusions: Studies included in this review are heterogeneous in study design, biological samples, and outcome. More coordinated investigations are needed to affirm DNA methylation as a clinically relevant biomarker important for prevention, monitoring, and intervention.
Keywords: Biomarker; DNA methylation; Kidney transplantation; Reperfusion injury; Systematic review.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Wolfe RA, et al. Comparison of mortality in all patients on dialysis, patients on dialysis awaiting transplantation, and recipients of a first cadaveric transplant. N Engl J Med. 1999;341(23):1725–1730. - PubMed
-
- Moreso F, Hernandez D. Has the survival of the graft improved after renal transplantation in the era of modern immunosuppression? Nefrología (English Edition) 2013;33(1):14–26. - PubMed
-
- Dupont PJ, Manuel O, Pascual M. Infection and chronic allograft dysfunction. Kidney Int Suppl. 2010;78(119):S47–S53. - PubMed
-
- Campistol JM, et al. Chronic allograft nephropathy: a clinical syndrome: early detection and the potential role of proliferation signal inhibitors. Clin Transplant. 2009;23(6):769–777. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

