Intravesical delivery of KDM6A-mRNA via mucoadhesive nanoparticles inhibits the metastasis of bladder cancer
- PMID: 35131941
- PMCID: PMC8851555
- DOI: 10.1073/pnas.2112696119
Intravesical delivery of KDM6A-mRNA via mucoadhesive nanoparticles inhibits the metastasis of bladder cancer
Abstract
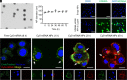
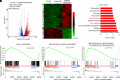
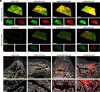
Lysine-specific demethylase 6A (KDM6A), also named UTX, is frequently mutated in bladder cancer (BCa). Although known as a tumor suppressor, KDM6A's therapeutic potential in the metastasis of BCa remains elusive. It also remains difficult to fulfill the effective up-regulation of KDM6A levels in bladder tumor tissues in situ to verify its potential in treating BCa metastasis. Here, we report a mucoadhesive messenger RNA (mRNA) nanoparticle (NP) strategy for the intravesical delivery of KDM6A-mRNA in mice bearing orthotopic Kdm6a-null BCa and show evidence of KDM6A's therapeutic potential in inhibiting the metastasis of BCa. Through this mucoadhesive mRNA NP strategy, the exposure of KDM6A-mRNA to the in situ BCa tumors can be greatly prolonged for effective expression, and the penetration can be also enhanced by adhering to the bladder for sustained delivery. This mRNA NP strategy is also demonstrated to be effective for combination cancer therapy with other clinically approved drugs (e.g., elemene), which could further enhance therapeutic outcomes. Our findings not only report intravesical delivery of mRNA via a mucoadhesive mRNA NP strategy but also provide the proof-of-concept for the usefulness of these mRNA NPs as tools in both mechanistic understanding and translational study of bladder-related diseases.
Keywords: KDM6A; bladder cancer; elemene; intravesical delivery; mRNA nanoparticles.
Copyright © 2022 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures



Comment in
-
Urolithiasis/Endourology.J Urol. 2022 Nov;208(5):1154-1155. doi: 10.1097/JU.0000000000002913. Epub 2022 Aug 17. J Urol. 2022. PMID: 35975568 No abstract available.
-
Uro-Science.J Urol. 2022 Nov;208(5):1152-1153. doi: 10.1097/JU.0000000000002932. Epub 2022 Aug 22. J Urol. 2022. PMID: 35993121 No abstract available.
References
-
- Ler L. D., et al. , Loss of tumor suppressor KDM6A amplifies PRC2-regulated transcriptional repression in bladder cancer and can be targeted through inhibition of EZH2. Sci. Transl. Med. 9, eaai8312 (2017). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

