Epigenetic study of early breast cancer (EBC) based on DNA methylation and gene integration analysis
- PMID: 35132081
- PMCID: PMC8821628
- DOI: 10.1038/s41598-022-05486-3
Epigenetic study of early breast cancer (EBC) based on DNA methylation and gene integration analysis
Erratum in
-
Author Correction: Epigenetic study of early breast cancer (EBC) based on DNA methylation and gene integration analysis.Sci Rep. 2022 May 26;12(1):8919. doi: 10.1038/s41598-022-13331-w. Sci Rep. 2022. PMID: 35618821 Free PMC article. No abstract available.
Abstract
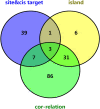
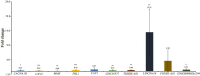
Breast cancer (BC) is one of the leading causes of cancer-related deaths in women. The purpose of this study is to identify key molecular markers related to the diagnosis and prognosis of early breast cancer (EBC). The data of mRNA, lncRNA and DNA methylation were downloaded from The Cancer Genome Atlas (TCGA) dataset for identification of differentially expressed mRNAs (DEmRNAs), differentially expressed lncRNAs (DElncRNAs) and DNA methylation analysis. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyzes were used to identify the biological functions of DEmRNAs. The correlation analysis between DNA methylation and DEmRNAs was carried out. Then, diagnostic analysis and prognostic analysis of identified DEmRNAs and DElncRNAs were also performed in the TCGA database. Subsequently, methylation state verification for identified DEmRNAs was performed in the GSE32393 dataset. In addition, real-time polymerase chain reaction (RT-PCR) in vitro verification of genes was performed. Finally, AC093110.1 was overexpressed in human BC cell line MCF-7 to verify cell proliferation and migration. In this study, a total of 1633 DEmRNAs, 750 DElncRNAs and 8042 differentially methylated sites were obtained, respectively. In the Venn analysis, 11 keys DEmRNAs (ALDH1L1, SPTBN1, MRGPRF, CAV2, HSPB6, PITX1, WDR86, PENK, CACNA1H, ALDH1A2 and MME) were we found. ALDH1A2, ALDH1L1, HSPB6, MME, MRGPRF, PENK, PITX1, SPTBN1, WDR86 and CAV2 may be considered as potential diagnostic gene biomarkers in EBC. Strikingly, CAV2, MME, AC093110.1 and AC120498.6 were significantly actively correlated with survival. Methylation state of identified DEmRNAs in GSE32393 dataset was consistent with the result in TCGA. AC093110.1 can affect the proliferation and migration of MCF-7. ALDH1A2, ALDH1L1, HSPB6, MME, MRGPRF, PENK, PITX1, SPTBN1, WDR86 and CAV2 may be potential diagnostic gene biomarkers of EBC. Strikingly, CAV2, MME, AC093110.1 and AC120498.6 were significantly actively correlated with survival. The identification of these genes can help in the early diagnosis and treatment of EBC. In addition, AC093110.1 can regulate SPTBN1 expression and play an important role in cell proliferation and migration, which provides clues to clarify the regulatory mechanism of EBC.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures










References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

