Consistency of variant interpretations among bioinformaticians and clinical geneticists in hereditary cancer panels
- PMID: 35132179
- PMCID: PMC8904571
- DOI: 10.1038/s41431-022-01060-7
Consistency of variant interpretations among bioinformaticians and clinical geneticists in hereditary cancer panels
Abstract
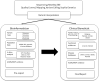
Next-generation sequencing (NGS) is used increasingly in hereditary cancer patients' (HCP) management. While enabling evaluation of multiple genes simultaneously, the technology brings to light the dilemma of variant interpretation. Here, we aimed to reveal the underlying reasons for the discrepancy in the evidence titles used during variant classification according to ACMG guidelines by two different bioinformatic specialists (BIs) and two different clinical geneticists (CGs). We evaluated final reports of 1920 cancer patients and 189 different variants from 285 HCP were enrolled to the study. A total of 173 of these variants were classified as pathogenic (n = 132) and likely pathogenic (n = 41) by the BI and an additional 16 variants, that were classified as VUS by at least one interpreter and their classification would change the clinical management, were compared for their evidence titles between different specialists. The attributed evidence titles and the final classification of the variants among BIs and CGs were compared. The discrepancy between P/LP final reports was 22.5%. The discordance between CGs was 30% whereas the discordance between two BIs was almost 75%. The use of PVS1, PS3, PP3, PP5, PM1, PM2, BP1, BP4 criteria markedly varied from one expert to another. This difference was particularly noticeable in PP3, PP5, and PM1 evidence and mostly in the variants affecting splice sites like BRCA1(NM_007294.4) c.4096 + 1 G > A and CHEK2(NM_007194.4) c.592 + 3 A > T. With recent advancements in precision medicine, the importance of variant interpretations is emerging. Our study shows that variant interpretation is subjective process that is in need of concrete definitions for accurate and standard interpretation.
© 2022. The Author(s), under exclusive licence to European Society of Human Genetics.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24.. doi: 10.1038/gim.2015.30. - DOI - PMC - PubMed
-
- National Comprehensive Cancer Network. Clinical practice guidelines in oncology genetic/familial high-risk assessment: colorectal. Available from: https://www.nccn.org/professionals/physician_gls/pdf/genetics_colon.pdf (2020). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

