NTR 2.0: a rationally engineered prodrug-converting enzyme with substantially enhanced efficacy for targeted cell ablation
- PMID: 35132245
- PMCID: PMC8851868
- DOI: 10.1038/s41592-021-01364-4
NTR 2.0: a rationally engineered prodrug-converting enzyme with substantially enhanced efficacy for targeted cell ablation
Abstract
Transgenic expression of bacterial nitroreductase (NTR) enzymes sensitizes eukaryotic cells to prodrugs such as metronidazole (MTZ), enabling selective cell-ablation paradigms that have expanded studies of cell function and regeneration in vertebrates. However, first-generation NTRs required confoundingly toxic prodrug treatments to achieve effective cell ablation, and some cell types have proven resistant. Here we used rational engineering and cross-species screening to develop an NTR variant, NTR 2.0, which exhibits ~100-fold improvement in MTZ-mediated cell-specific ablation efficacy, eliminating the need for near-toxic prodrug treatment regimens. NTR 2.0 therefore enables sustained cell-loss paradigms and ablation of previously resistant cell types. These properties permit enhanced interrogations of cell function, extended challenges to the regenerative capacities of discrete stem cell niches, and novel modeling of chronic degenerative diseases. Accordingly, we have created a series of bipartite transgenic reporter/effector resources to facilitate dissemination of NTR 2.0 to the research community.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing Interests Statement
J.S.M. has been awarded patents for the creation (US patent #7,514,595) and use of zebrafish expressing nitroreductase enzymes for gene (US patent #8,071,838) and drug discovery (US patent #8,431,768) applications. M.T.S. is the President and Scientific Director at Luminomics, a biotechnology start-up that offers ARQiv-based screening services. M.T.S. owns stock in Luminomics and J.S.M. serves as a consultant at Luminomics. The remaining authors declare no competing interests.
Figures

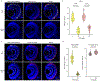

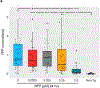



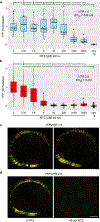
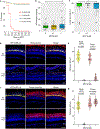

References
-
- Williams EM et al. Nitroreductase gene-directed enzyme prodrug therapy: insights and advances toward clinical utility. Biochem. J 471, 131–153 (2015). - PubMed
-
- Roldán MD, Pérez-Reinado E, Castillo F & Moreno-Vivián C Reduction of polynitroaromatic compounds: the bacterial nitroreductases. FEMS Microbiol. Rev 32, 474–500 (2008). - PubMed
-
- Venitt S & Crofton-Sleigh C The toxicity and mutagenicity of the anti-tumour drug 5-aziridino-2,4-dinitrobenzamide (CB1954) is greatly reduced in a nitroreductase-deficient strain of E. coli. Mutagenesis 2, 375–81 (1987). - PubMed
-
- Knox RJ et al. The nitroreductase enzyme in Walker cells that activates 5-(aziridin-1-yl)-2,4-dinitrobenzamide (CB 1954) to 5-(aziridin-1-yl)-4-hydroxylamino-2-nitrobenzamide is a form of NAD(P)H dehydrogenase (quinone) (EC 1.6.99.2). Biochem. Pharmacol 37, 4671–7 (1988). - PubMed
-
- Lewis K Platforms for antibiotic discovery. Nat. Rev. Drug Discov 12, 371–387 (2013). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

