HNF4A Regulates the Proliferation and Tumor Formation of Cervical Cancer Cells through the Wnt/ β-Catenin Pathway
- PMID: 35132353
- PMCID: PMC8817108
- DOI: 10.1155/2022/8168988
HNF4A Regulates the Proliferation and Tumor Formation of Cervical Cancer Cells through the Wnt/ β-Catenin Pathway
Abstract
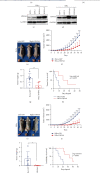
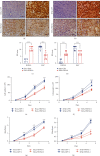
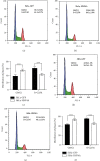
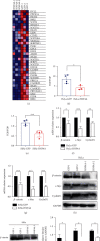
Hepatocyte nuclear factor 4 alpha (HNF4A) is a transcriptional factor which plays an important role in the development of the liver, kidney, and intestines. Nevertheless, its role in cervical cancer and the underlying mechanism remain unknown. In this study, both immunohistochemistry and western blotting revealed that the expression of HNF4A was downregulated in cervical cancer. Xenograft assays suggested that HN4A could inhibit tumorigenic potential of cervical cancer in vivo. Functional studies illustrated that HNF4A also inhibited the proliferation and viability of cervical cancer cells in vitro. In addition, FACS analysis implied that HNF4A could induce cell cycle arrest from the G0/G1 phase to S phase. Further studies suggested that HNF4A downregulated the activity of the Wnt/β-catenin pathway. Altogether, our data demonstrated that HNF4A inhibited tumor formation and proliferation of cervical cancer cells through suppressing the activity of the Wnt/β-catenin pathway.
Copyright © 2022 Hong-Mei Ma et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures

Similar articles
-
SOX17 restrains proliferation and tumor formation by down-regulating activity of the Wnt/β-catenin signaling pathway via trans-suppressing β-catenin in cervical cancer.Cell Death Dis. 2018 Jul 3;9(7):741. doi: 10.1038/s41419-018-0782-8. Cell Death Dis. 2018. PMID: 29970906 Free PMC article.
-
HOXA5 inhibits the proliferation and neoplasia of cervical cancer cells via downregulating the activity of the Wnt/β-catenin pathway and transactivating TP53.Cell Death Dis. 2020 Jun 4;11(6):420. doi: 10.1038/s41419-020-2629-3. Cell Death Dis. 2020. PMID: 32499530 Free PMC article.
-
SALL4 promotes the tumorigenicity of cervical cancer cells through activation of the Wnt/β-catenin pathway via CTNNB1.Cancer Sci. 2019 Sep;110(9):2794-2805. doi: 10.1111/cas.14140. Epub 2019 Aug 16. Cancer Sci. 2019. PMID: 31336010 Free PMC article.
-
EZH2-mediated repression of GSK-3β and TP53 promotes Wnt/β-catenin signaling-dependent cell expansion in cervical carcinoma.Oncotarget. 2016 Jun 14;7(24):36115-36129. doi: 10.18632/oncotarget.8741. Oncotarget. 2016. PMID: 27092879 Free PMC article.
-
HOXB4 inhibits the proliferation and tumorigenesis of cervical cancer cells by downregulating the activity of Wnt/β-catenin signaling pathway.Cell Death Dis. 2021 Jan 21;12(1):105. doi: 10.1038/s41419-021-03411-6. Cell Death Dis. 2021. PMID: 33479226 Free PMC article.
Cited by
-
SNHG3/WISP2 Axis Promotes Hela Cell Migration and Invasion via Activating Wnt/β-Catenin Signaling.Cancer Genomics Proteomics. 2023 Dec;20(6suppl):744-753. doi: 10.21873/cgp.20421. Cancer Genomics Proteomics. 2023. PMID: 38035707 Free PMC article.
-
HNF4A promotes tumor progression in gastric cancer by transcriptional activation of KIF2C.Clin Exp Med. 2025 Jun 14;25(1):205. doi: 10.1007/s10238-025-01748-2. Clin Exp Med. 2025. PMID: 40517212 Free PMC article.
-
Unveiling the link between arsenic toxicity and diabetes: an in silico exploration into the role of transcription factors.Toxicol Res. 2024 Jul 18;40(4):653-672. doi: 10.1007/s43188-024-00255-y. eCollection 2024 Oct. Toxicol Res. 2024. PMID: 39345741
-
Sex-Specific Concordance of Striatal Transcriptional Signatures of Opioid Addiction in Human and Rodent Brains.Biol Psychiatry Glob Open Sci. 2025 Feb 26;5(3):100476. doi: 10.1016/j.bpsgos.2025.100476. eCollection 2025 May. Biol Psychiatry Glob Open Sci. 2025. PMID: 40248277 Free PMC article.
-
Orphan nuclear receptor transcription factors as drug targets.Transcription. 2025 Apr-Jun;16(2-3):224-260. doi: 10.1080/21541264.2025.2521766. Epub 2025 Jul 11. Transcription. 2025. PMID: 40646688 Free PMC article. Review.
References
-
- Bray F., Ferlay J., Soerjomataram I., Siegel R. L., Torre L. A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a Cancer Journal for Clinicians . 2018;68(6):394–424. - PubMed
-
- Mighty K. K., Laimins L. A. The role of human papillomaviruses in oncogenesis. Recent results in cancer research Fortschritte der Krebsforschung Progres dans les recherches sur le cancer . 2014;193:135–148. - PubMed

