This is a preprint.
A Model for Network-Based Identification and Pharmacological Targeting of Aberrant, Replication-Permissive Transcriptional Programs Induced by Viral Infection
- PMID: 35132404
- PMCID: PMC8820669
- DOI: 10.21203/rs.3.rs-1287631/v1
A Model for Network-Based Identification and Pharmacological Targeting of Aberrant, Replication-Permissive Transcriptional Programs Induced by Viral Infection
Update in
-
A model for network-based identification and pharmacological targeting of aberrant, replication-permissive transcriptional programs induced by viral infection.Commun Biol. 2022 Jul 19;5(1):714. doi: 10.1038/s42003-022-03663-8. Commun Biol. 2022. PMID: 35854100 Free PMC article.
Abstract
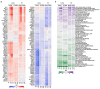
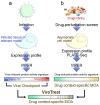
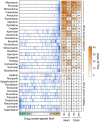
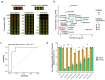
Precise characterization and targeting of host cell transcriptional machinery hijacked by viral infection remains challenging. Here, we show that SARS-CoV-2 hijacks the host cell transcriptional machinery to induce a phenotypic state amenable to its replication. Specifically, analysis of Master Regulator (MR) proteins representing mechanistic determinants of the gene expression signature induced by SARS-CoV-2 in infected cells revealed coordinated inactivation of MRs enriched in physical interactions with SARS-CoV-2 proteins, suggesting their mechanistic role in maintaining a host cell state refractory to virus replication. To test their functional relevance, we measured SARS-CoV-2 replication in epithelial cells treated with drugs predicted to activate the entire repertoire of repressed MRs, based on their experimentally elucidated, context-specific mechanism of action. Overall, >80% of drugs predicted to be effective by this methodology induced significant reduction of SARS-CoV-2 replication, without affecting cell viability. This model for host-directed pharmacological therapy is fully generalizable and can be deployed to identify drugs targeting host cell-based MR signatures induced by virtually any pathogen.
Conflict of interest statement
Figures
Similar articles
-
A model for network-based identification and pharmacological targeting of aberrant, replication-permissive transcriptional programs induced by viral infection.Commun Biol. 2022 Jul 19;5(1):714. doi: 10.1038/s42003-022-03663-8. Commun Biol. 2022. PMID: 35854100 Free PMC article.
-
The Host Cell ViroCheckpoint: Identification and Pharmacologic Targeting of Novel Mechanistic Determinants of Coronavirus-Mediated Hijacked Cell States.bioRxiv [Preprint]. 2020 May 17:2020.05.12.091256. doi: 10.1101/2020.05.12.091256. bioRxiv. 2020. PMID: 32511361 Free PMC article. Preprint.
-
Identification and Targeting of Regulators of SARS-CoV-2-Host Interactions in the Airway Epithelium.bioRxiv [Preprint]. 2024 Dec 9:2024.10.11.617898. doi: 10.1101/2024.10.11.617898. bioRxiv. 2024. Update in: Sci Adv. 2025 May 16;11(20):eadu2079. doi: 10.1126/sciadv.adu2079. PMID: 39464067 Free PMC article. Updated. Preprint.
-
Bis(monoacylglycero)phosphate, an important actor in the host endocytic machinery hijacked by SARS-CoV-2 and related viruses.Biochimie. 2020 Dec;179:247-256. doi: 10.1016/j.biochi.2020.10.018. Epub 2020 Nov 5. Biochimie. 2020. PMID: 33159981 Free PMC article. Review.
-
Myocardial Damage by SARS-CoV-2: Emerging Mechanisms and Therapies.Viruses. 2021 Sep 21;13(9):1880. doi: 10.3390/v13091880. Viruses. 2021. PMID: 34578462 Free PMC article. Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

