Spatiotemporal Analyses of 2 Co-Circulating SARS-CoV-2 Variants, New York State, USA
- PMID: 35133957
- PMCID: PMC8888210
- DOI: 10.3201/eid2803.211972
Spatiotemporal Analyses of 2 Co-Circulating SARS-CoV-2 Variants, New York State, USA
Abstract
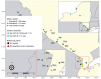
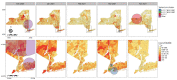
The emergence of novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants in late 2020 and early 2021 raised alarm worldwide because of their potential for increased transmissibility and immune evasion. Elucidating the evolutionary and epidemiologic dynamics among novel SARS-CoV-2 variants is essential for understanding the trajectory of the coronavirus disease pandemic. We describe the interplay between B.1.1.7 (Alpha) and B.1.526 (Iota) variants in New York State, USA, during December 2020-April 2021 through phylogeographic analyses, space-time scan statistics, and cartographic visualization. Our results indicate that B.1.526 probably evolved in New York City, where it was displaced as the dominant lineage by B.1.1.7 months after its initial appearance. In contrast, B.1.1.7 became dominant earlier in regions with fewer B.1.526 infections. These results suggest that B.1.526 might have delayed the initial spread of B.1.1.7 in New York City. Our combined spatiotemporal methodologies can help disentangle the complexities of shifting SARS-CoV-2 variant landscapes.
Keywords: COVID-19; New York; SARS-CoV-2; United States; coronavirus disease; respiratory infections; severe acute respiratory syndrome coronavirus 2; spatiotemporal analyses; vaccine-preventable diseases; viruses; zoonoses.
Figures


References
-
- Frampton D, Rampling T, Cross A, Bailey H, Heaney J, Byott M, et al. Genomic characteristics and clinical effect of the emergent SARS-CoV-2 B.1.1.7 lineage in London, UK: a whole-genome sequencing and hospital-based cohort study. Lancet Infect Dis. 2021;21:1246–56. 10.1016/S1473-3099(21)00170-5 - DOI - PMC - PubMed
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

