Recruitment and training of alveolar macrophages after pneumococcal pneumonia
- PMID: 35133985
- PMCID: PMC8983128
- DOI: 10.1172/jci.insight.150239
Recruitment and training of alveolar macrophages after pneumococcal pneumonia
Abstract
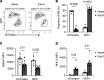
Recovery from pneumococcal pneumonia remodels the pool of alveolar macrophages so that they exhibit new surface marker profiles, transcriptomes, metabolomes, and responses to infection. Mechanisms mediating alveolar macrophage phenotypes after pneumococcal pneumonia have not been delineated. IFN-γ and its receptor on alveolar macrophages were essential for certain, but not all, aspects of the remodeled alveolar macrophage phenotype. IFN-γ was produced by CD4+ T cells plus other cells, and CD4+ cell depletion did not prevent alveolar macrophage remodeling. In mice infected or recovering from pneumococcus, monocytes were recruited to the lungs, and the monocyte-derived macrophages developed characteristics of alveolar macrophages. CCR2 mediated the early monocyte recruitment but was not essential to the development of the remodeled alveolar macrophage phenotype. Lineage tracing demonstrated that recovery from pneumococcal pneumonias converted the pool of alveolar macrophages from being primarily of embryonic origin to being primarily of adult hematopoietic stem cell origin. Alveolar macrophages of either origin demonstrated similar remodeled phenotypes, suggesting that ontogeny did not dictate phenotype. Our data reveal that the remodeled alveolar macrophage phenotype in lungs recovered from pneumococcal pneumonia results from a combination of new recruitment plus training of both the original cells and the new recruits.
Keywords: Bacterial infections; Immunology; Macrophages; Pulmonology.
Conflict of interest statement
Figures

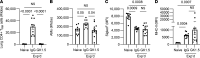








References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

