Heterochiasmy and the establishment of gsdf as a novel sex determining gene in Atlantic halibut
- PMID: 35134055
- PMCID: PMC8824383
- DOI: 10.1371/journal.pgen.1010011
Heterochiasmy and the establishment of gsdf as a novel sex determining gene in Atlantic halibut
Abstract
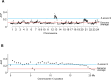
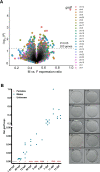
Atlantic Halibut (Hippoglossus hippoglossus) has a X/Y genetic sex determination system, but the sex determining factor is not known. We produced a high-quality genome assembly from a male and identified parts of chromosome 13 as the Y chromosome due to sequence divergence between sexes and segregation of sex genotypes in pedigrees. Linkage analysis revealed that all chromosomes exhibit heterochiasmy, i.e. male-only and female-only meiotic recombination regions (MRR/FRR). We show that FRR/MRR intervals differ in nucleotide diversity and repeat class content and that this is true also for other Pleuronectidae species. We further show that remnants of a Gypsy-like transposable element insertion on chr13 promotes early male specific expression of gonadal somatic cell derived factor (gsdf). Less than 4.5 MYA, this male-determining element evolved on an autosomal FRR segment featuring pre-existing male meiotic recombination barriers, thereby creating a Y chromosome. Our findings indicate that heterochiasmy may facilitate the evolution of genetic sex determination systems relying on linkage of sexually antagonistic loci to a sex-determining factor.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

