Sequencing SARS-CoV-2 from antigen tests
- PMID: 35134077
- PMCID: PMC8824375
- DOI: 10.1371/journal.pone.0263794
Sequencing SARS-CoV-2 from antigen tests
Abstract
Genomic surveillance empowers agile responses to SARS-CoV-2 by enabling scientists and public health analysts to issue recommendations aimed at slowing transmission, prioritizing contact tracing, and building a robust genomic sequencing surveillance strategy. Since the start of the pandemic, real time RT-PCR diagnostic testing from upper respiratory specimens, such as nasopharyngeal (NP) swabs, has been the standard. Moreover, respiratory samples in viral transport media are the ideal specimen for SARS-CoV-2 whole-genome sequencing (WGS). In early 2021, many clinicians transitioned to antigen-based SARS-CoV-2 detection tests, which use anterior nasal swabs for SARS-CoV-2 antigen detection. Despite this shift in testing methods, the need for whole-genome sequence surveillance remains. Thus, we developed a workflow for whole-genome sequencing with antigen test-derived swabs as an input rather than nasopharyngeal swabs. In this study, we use excess clinical specimens processed using the BinaxNOW™ COVID-19 Ag Card. We demonstrate that whole-genome sequencing from antigen tests is feasible and yields similar results from RT-PCR-based assays utilizing a swab in viral transport media.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
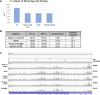
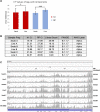
References
-
- Long SW, Olsen RJ, Christensen PA, Subedi S, Olson R, Davis JJ, et al.. Sequence Analysis of 20,453 Severe Acute Respiratory Syndrome Coronavirus 2 Genomes from the Houston Metropolitan Area Identifies the Emergence and Widespread Distribution of Multiple Isolates of All Major Variants of Concern. Am J Pathol. 2021;191: 983–992. doi: 10.1016/j.ajpath.2021.03.004 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

