Chemokine C-X-C receptor 4 mediates recruitment of bone marrow-derived nonhematopoietic and immune cells to the pregnant uterus†
- PMID: 35134114
- PMCID: PMC9198949
- DOI: 10.1093/biolre/ioac029
Chemokine C-X-C receptor 4 mediates recruitment of bone marrow-derived nonhematopoietic and immune cells to the pregnant uterus†
Abstract
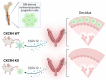
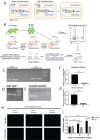
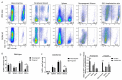
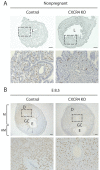
Bone marrow-derived progenitor cells (BMDPCs) are mobilized to the circulation in pregnancy and get recruited to the pregnant decidua where they contribute functionally to decidualization and successful implantation. However, the molecular mechanisms underlying BMDPCs recruitment to the decidua are unknown. CXCL12 ligand and its CXCR4 receptor play crucial roles in the mobilization and homing of stem/progenitor cells to various tissues. To investigate the role of CXCL12-CXCR4 axis in BMDPCs recruitment to decidua, we created transgenic GFP mice harboring CXCR4 gene susceptible to tamoxifen-inducible Cre-mediated ablation. These mice served as BM donors into wild-type C57BL/6 J female recipients using a 5-fluorouracil-based nongonadotoxic submyeloablation to achieve BM-specific CXCR4 knockout (CXCR4KO). Successful CXCR4 ablation was confirmed by RT-PCR and in vitro cell migration assays. Flow cytometry and immunohistochemistry showed a significant increase in GFP+ BM-derived cells (BMDCs) in the implantation site as compared to the nonpregnant uterus of control (2.7-fold) and CXCR4KO (1.8-fold) mice. This increase was uterus-specific and was not observed in other organs. This pregnancy-induced increase occurred in both hematopoietic (CD45+) and nonhematopoietic (CD45-) uterine BMDCs in control mice. In contrast, in CXCR4KO mice there was no increase in nonhematopoietic BMDCs in the pregnant uterus. Moreover, decidual recruitment of myeloid cells but not NK cells was diminished by BM CXCR4 deletion. Immunofluorescence showed the presence of nonhematopoietic GFP+ cells that were negative for CD45 (panleukocyte) and DBA (NK) markers in control but not CXCR4KO decidua. In conclusion, we report that CXCR4 expression in nonhematopoietic BMDPCs is essential for their recruitment to the pregnant decidua.
Keywords: CXCR4; bone marrow; implantation; pregnancy; stem cells; uterus.
© The Author(s) 2022. Published by Oxford University Press on behalf of Society for the Study of Reproduction. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures




Similar articles
-
Bone-marrow-derived endothelial progenitor cells contribute to vasculogenesis of pregnant mouse uterus†.Biol Reprod. 2019 May 1;100(5):1228-1237. doi: 10.1093/biolre/ioy265. Biol Reprod. 2019. PMID: 30601943 Free PMC article.
-
Adult bone marrow progenitors become decidual cells and contribute to embryo implantation and pregnancy.PLoS Biol. 2019 Sep 12;17(9):e3000421. doi: 10.1371/journal.pbio.3000421. eCollection 2019 Sep. PLoS Biol. 2019. PMID: 31513564 Free PMC article.
-
Bone marrow-derived progenitor cells contribute to remodeling of the postpartum uterus.Stem Cells. 2021 Nov;39(11):1489-1505. doi: 10.1002/stem.3431. Epub 2021 Aug 6. Stem Cells. 2021. PMID: 34224633 Free PMC article.
-
Mutual, reciprocal SDF-1/CXCR4 interactions between hematopoietic and bone marrow stromal cells regulate human stem cell migration and development in NOD/SCID chimeric mice.Exp Hematol. 2006 Aug;34(8):967-75. doi: 10.1016/j.exphem.2006.04.002. Exp Hematol. 2006. PMID: 16863903 Review.
-
Innate immunity derived factors as external modulators of the CXCL12-CXCR4 axis and their role in stem cell homing and mobilization.Theranostics. 2013;3(1):3-10. doi: 10.7150/thno.4621. Epub 2013 Jan 12. Theranostics. 2013. PMID: 23382780 Free PMC article. Review.
Cited by
-
Uterus: A Unique Stem Cell Reservoir Able to Support Cardiac Repair via Crosstalk among Uterus, Heart, and Bone Marrow.Cells. 2022 Jul 13;11(14):2182. doi: 10.3390/cells11142182. Cells. 2022. PMID: 35883625 Free PMC article. Review.
-
Maternal CXCR4 deletion results in placental defects and pregnancy loss mediated by immune dysregulation.JCI Insight. 2023 Nov 8;8(21):e172216. doi: 10.1172/jci.insight.172216. JCI Insight. 2023. PMID: 37815869 Free PMC article.
-
Estimate the relationship between CXCR4-SDF-1 axis and inhibitory molecules (CTLA4 and PD-1) in patients with colon cancer.Narra J. 2024 Dec;4(3):e992. doi: 10.52225/narra.v4i3.992. Epub 2024 Dec 16. Narra J. 2024. PMID: 39816054 Free PMC article.
References
-
- Fievez V, Szpakowska M, Mosbah A, Arumugam K, Mathu J, Counson M, Beaupain N, Seguin-Devaux C, Deroo S, Baudy-Floc'h M, Chevigné A. Development of mimokines, chemokine N terminus-based CXCR4 inhibitors optimized by phage display and rational design. J Leukoc Biol 2018; 104:343–357. - PubMed
-
- Karpova D, Bonig H. Concise review: CXCR4/CXCL12 signaling in immature hematopoiesis--lessons from pharmacological and genetic models. Stem Cells 2015; 33:2391–2399. - PubMed
-
- Sugiyama T, Kohara H, Noda M, Nagasawa T. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity 2006; 25:977–988. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

