Unbiased cell surface proteomics identifies SEMA4A as an effective immunotherapy target for myeloma
- PMID: 35134130
- PMCID: PMC11022854
- DOI: 10.1182/blood.2021015161
Unbiased cell surface proteomics identifies SEMA4A as an effective immunotherapy target for myeloma
Abstract
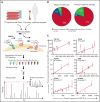
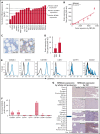
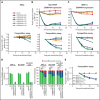
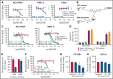
The accessibility of cell surface proteins makes them tractable for targeting by cancer immunotherapy, but identifying suitable targets remains challenging. Here we describe plasma membrane profiling of primary human myeloma cells to identify an unprecedented number of cell surface proteins of a primary cancer. We used a novel approach to prioritize immunotherapy targets and identified a cell surface protein not previously implicated in myeloma, semaphorin-4A (SEMA4A). Using knock-down by short-hairpin RNA and CRISPR/nuclease-dead Cas9 (dCas9), we show that expression of SEMA4A is essential for normal myeloma cell growth in vitro, indicating that myeloma cells cannot downregulate the protein to avoid detection. We further show that SEMA4A would not be identified as a myeloma therapeutic target by standard CRISPR/Cas9 knockout screens because of exon skipping. Finally, we potently and selectively targeted SEMA4A with a novel antibody-drug conjugate in vitro and in vivo.
© 2022 by The American Society of Hematology.
Conflict of interest statement
Figures



Comment in
-
New immunotherapeutic target in myeloma.Blood. 2022 Apr 21;139(16):2417-2418. doi: 10.1182/blood.2022015481. Blood. 2022. PMID: 35446380 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

