Tracing the SARS-CoV-2 infection on the ocular surface: Overview and preliminary corneoscleral transcriptome sequencing
- PMID: 35134391
- PMCID: PMC8816849
- DOI: 10.1016/j.exer.2022.108975
Tracing the SARS-CoV-2 infection on the ocular surface: Overview and preliminary corneoscleral transcriptome sequencing
Abstract
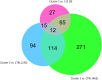
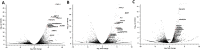
COVID-19's impact on the ocular surface has already been recognized, however the molecular mechanisms induced by the infection on the ocular surface are still unclear. The aim of this paper is to provide a first overview of the transcriptional perturbations caused by SARS-CoV-2 on the ocular surface by analyzing gene expression profile of corneoscleral ring samples from post-mortem SARS-CoV-2 positive donors (PD). The presence of SARS-CoV-2 on the ocular surface, in tears and corneal tissues has rarely been detected in infected individuals in both the presence and the absence of ocular manifestations. In this preliminary study, 6 human corneoscleral tissues of 3 PD and two tissues from a negative donor (CTRL) were obtained at the local eye bank. The presence of genomic and sub-genomic SARS-CoV-2 RNAs was assessed by qRT-PCR, while transcriptome analysis (RNA-sequencing) was performed by Illumina. Principal Component Analysis (PCA), search for differentially expressed genes (DEGs) and Gene Ontology (GO)-enrichment analysis were performed. Three samples from PD were found positive for SARS-CoV-2 genomic RNA, although the absence of sub-genomic RNAs indicated an inactive virus. PCA analysis grouped 3 different clusters, one including CTRL, and the other two including, respectively, PD with undetected SARS-CoV-2 (PD-SARS-neg) and PD with detected SARS-CoV-2 (PD-SARS-pos). The DEGs in common with the 2 PD clusters included several genes associable to the interferon pathway, such as ADAMTS4, RSAD2, MMP1, IL6, ISG15 and proinflammatory cytokines. Among the down-regulated genes we found AQP5. GO analysis revealed 77 GO terms over-represented in PD-SARS-neg vs. CTRL, and 17 GO terms in PD-SARS-pos vs. CTRL. The presence of SARS-CoV-2 RNA and RNA-sequencing reads in ocular surface tissues supports the possibility that the eye acts as an entry route. The modulation of early responsive genes, together with several ISGs suggests a potential protective responsiveness of the ocular tissues to SARS-CoV-2.
Keywords: Cornea; Corneoscleral ring; Ocular surface; RNA-Sequencing; SARS-CoV-2.
Copyright © 2022 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Authors have no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

