A deep dilated convolutional residual network for predicting interchain contacts of protein homodimers
- PMID: 35134816
- PMCID: PMC8963319
- DOI: 10.1093/bioinformatics/btac063
A deep dilated convolutional residual network for predicting interchain contacts of protein homodimers
Abstract
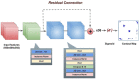
Motivation: Deep learning has revolutionized protein tertiary structure prediction recently. The cutting-edge deep learning methods such as AlphaFold can predict high-accuracy tertiary structures for most individual protein chains. However, the accuracy of predicting quaternary structures of protein complexes consisting of multiple chains is still relatively low due to lack of advanced deep learning methods in the field. Because interchain residue-residue contacts can be used as distance restraints to guide quaternary structure modeling, here we develop a deep dilated convolutional residual network method (DRCon) to predict interchain residue-residue contacts in homodimers from residue-residue co-evolutionary signals derived from multiple sequence alignments of monomers, intrachain residue-residue contacts of monomers extracted from true/predicted tertiary structures or predicted by deep learning, and other sequence and structural features.
Results: Tested on three homodimer test datasets (Homo_std dataset, DeepHomo dataset and CASP-CAPRI dataset), the precision of DRCon for top L/5 interchain contact predictions (L: length of monomer in a homodimer) is 43.46%, 47.10% and 33.50% respectively at 6 Å contact threshold, which is substantially better than DeepHomo and DNCON2_inter and similar to Glinter. Moreover, our experiments demonstrate that using predicted tertiary structure or intrachain contacts of monomers in the unbound state as input, DRCon still performs well, even though its accuracy is lower than using true tertiary structures in the bound state are used as input. Finally, our case study shows that good interchain contact predictions can be used to build high-accuracy quaternary structure models of homodimers.
Availability and implementation: The source code of DRCon is available at https://github.com/jianlin-cheng/DRCon. The datasets are available at https://zenodo.org/record/5998532#.YgF70vXMKsB.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2022. Published by Oxford University Press.
Figures



Similar articles
-
DNCON2_Inter: predicting interchain contacts for homodimeric and homomultimeric protein complexes using multiple sequence alignments of monomers and deep learning.Sci Rep. 2021 Jun 10;11(1):12295. doi: 10.1038/s41598-021-91827-7. Sci Rep. 2021. PMID: 34112907 Free PMC article.
-
Deep graph learning of inter-protein contacts.Bioinformatics. 2022 Jan 27;38(4):947-953. doi: 10.1093/bioinformatics/btab761. Bioinformatics. 2022. PMID: 34755837 Free PMC article.
-
DNCON2: improved protein contact prediction using two-level deep convolutional neural networks.Bioinformatics. 2018 May 1;34(9):1466-1472. doi: 10.1093/bioinformatics/btx781. Bioinformatics. 2018. PMID: 29228185 Free PMC article.
-
Recent Applications of Deep Learning Methods on Evolution- and Contact-Based Protein Structure Prediction.Int J Mol Sci. 2021 Jun 2;22(11):6032. doi: 10.3390/ijms22116032. Int J Mol Sci. 2021. PMID: 34199677 Free PMC article. Review.
-
The trRosetta server for fast and accurate protein structure prediction.Nat Protoc. 2021 Dec;16(12):5634-5651. doi: 10.1038/s41596-021-00628-9. Epub 2021 Nov 10. Nat Protoc. 2021. PMID: 34759384 Review.
Cited by
-
Flexible protein-protein docking with a multitrack iterative transformer.Protein Sci. 2024 Feb;33(2):e4862. doi: 10.1002/pro.4862. Protein Sci. 2024. PMID: 38148272 Free PMC article.
-
Combining pairwise structural similarity and deep learning interface contact prediction to estimate protein complex model accuracy in CASP15.Proteins. 2023 Dec;91(12):1889-1902. doi: 10.1002/prot.26542. Epub 2023 Jun 26. Proteins. 2023. PMID: 37357816 Free PMC article.
-
Noninvasive detection and interpretation of gastrointestinal diseases by collaborative serum metabolite and magnetically controlled capsule endoscopy.Comput Struct Biotechnol J. 2022 Oct 6;20:5524-5534. doi: 10.1016/j.csbj.2022.10.001. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 36249561 Free PMC article.
-
DataDTA: a multi-feature and dual-interaction aggregation framework for drug-target binding affinity prediction.Bioinformatics. 2023 Sep 2;39(9):btad560. doi: 10.1093/bioinformatics/btad560. Bioinformatics. 2023. PMID: 37688568 Free PMC article.
-
Prediction of inter-chain distance maps of protein complexes with 2D attention-based deep neural networks.Nat Commun. 2022 Nov 15;13(1):6963. doi: 10.1038/s41467-022-34600-2. Nat Commun. 2022. PMID: 36379943 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

