Vivarium: an interface and engine for integrative multiscale modeling in computational biology
- PMID: 35134830
- PMCID: PMC8963310
- DOI: 10.1093/bioinformatics/btac049
Vivarium: an interface and engine for integrative multiscale modeling in computational biology
Abstract
Motivation: This article introduces Vivarium-software born of the idea that it should be as easy as possible for computational biologists to define any imaginable mechanistic model, combine it with existing models and execute them together as an integrated multiscale model. Integrative multiscale modeling confronts the complexity of biology by combining heterogeneous datasets and diverse modeling strategies into unified representations. These integrated models are then run to simulate how the hypothesized mechanisms operate as a whole. But building such models has been a labor-intensive process that requires many contributors, and they are still primarily developed on a case-by-case basis with each project starting anew. New software tools that streamline the integrative modeling effort and facilitate collaboration are therefore essential for future computational biologists.
Results: Vivarium is a software tool for building integrative multiscale models. It provides an interface that makes individual models into modules that can be wired together in large composite models, parallelized across multiple CPUs and run with Vivarium's discrete-event simulation engine. Vivarium's utility is demonstrated by building composite models that combine several modeling frameworks: agent-based models, ordinary differential equations, stochastic reaction systems, constraint-based models, solid-body physics and spatial diffusion. This demonstrates just the beginning of what is possible-Vivarium will be able to support future efforts that integrate many more types of models and at many more biological scales.
Availability and implementation: The specific models, simulation pipelines and notebooks developed for this article are all available at the vivarium-notebooks repository: https://github.com/vivarium-collective/vivarium-notebooks. Vivarium-core is available at https://github.com/vivarium-collective/vivarium-core, and has been released on Python Package Index. The Vivarium Collective (https://vivarium-collective.github.io) is a repository of freely available Vivarium processes and composites, including the processes used in Section 3. Supplementary Materials provide with an extensive methodology section, with several code listings that demonstrate the basic interfaces.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2022. Published by Oxford University Press. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures
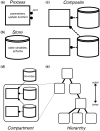
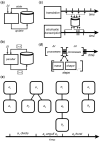

References
-
- Arjunan S., Tomita M. (2009) Modeling reaction-diffusion of molecules on surface and in volume spaces with the E-Cell system. Nat. Precedings, 1–1.
-
- Bartley B. et al. (2015) Synthetic biology open language (SBOL) version 2.0. 0. J. Integrative Bioinf., 12, 902–991. - PubMed

