Interpretable network-guided epistasis detection
- PMID: 35134928
- PMCID: PMC8848319
- DOI: 10.1093/gigascience/giab093
Interpretable network-guided epistasis detection
Abstract
Background: Detecting epistatic interactions at the gene level is essential to understanding the biological mechanisms of complex diseases. Unfortunately, genome-wide interaction association studies involve many statistical challenges that make such detection hard. We propose a multi-step protocol for epistasis detection along the edges of a gene-gene co-function network. Such an approach reduces the number of tests performed and provides interpretable interactions while keeping type I error controlled. Yet, mapping gene interactions into testable single-nucleotide polymorphism (SNP)-interaction hypotheses, as well as computing gene pair association scores from SNP pair ones, is not trivial.
Results: Here we compare 3 SNP-gene mappings (positional overlap, expression quantitative trait loci, and proximity in 3D structure) and use the adaptive truncated product method to compute gene pair scores. This method is non-parametric, does not require a known null distribution, and is fast to compute. We apply multiple variants of this protocol to a genome-wide association study dataset on inflammatory bowel disease. Different configurations produced different results, highlighting that various mechanisms are implicated in inflammatory bowel disease, while at the same time, results overlapped with known disease characteristics. Importantly, the proposed pipeline also differs from a conventional approach where no network is used, showing the potential for additional discoveries when prior biological knowledge is incorporated into epistasis detection.
Keywords: biology-informed analysis; epistasis network; gene-gene interaction; inflammatory bowel disease; systems biology.
© The Author(s) 2022. Published by Oxford University Press GigaScience.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

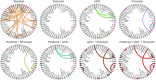



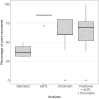

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

