Trade-offs between cost of ingestion and rate of intake drive defensive toxin use
- PMID: 35135316
- PMCID: PMC8826133
- DOI: 10.1098/rsbl.2021.0579
Trade-offs between cost of ingestion and rate of intake drive defensive toxin use
Abstract
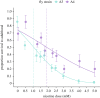
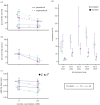
Animals that ingest toxins can become unpalatable and even toxic to predators and parasites through toxin sequestration. Because most animals rapidly eliminate toxins to survive their ingestion, it is unclear how populations transition from susceptibility and toxin elimination to tolerance and accumulation as chemical defence emerges. Studies of chemical defence have generally focused on species with active toxin sequestration and target-site insensitivity mutations or toxin-binding proteins that permit survival without necessitating toxin elimination. Here, we investigate whether animals that presumably rely on toxin elimination for survival can use ingested toxins for defence. We use the A4 and A3 Drosophila melanogaster fly strains from the Drosophila Synthetic Population Resource (DSPR), which respectively possess high and low metabolic nicotine resistance among DSPR fly lines. We find that ingesting nicotine increased A4 but not A3 fly survival against Leptopilina heterotoma wasp parasitism. Further, we find that despite possessing genetic variants that enhance toxin elimination, A4 flies accrued more nicotine than A3 individuals, likely by consuming more medium. Our results suggest that enhanced toxin metabolism can allow greater toxin intake by offsetting the cost of toxin ingestion. Passive toxin accumulation that accompanies increased toxin intake may underlie the early origins of chemical defence.
Keywords: bioaccumulation; chemical defence; enemy-free space; multi-trophic selection; xenobiotic metabolism.
Figures
References
-
- Duffey SS. 1980. Sequestration of plant natural products by insects. Annu. Rev. Entomol. 25, 447-477. (10.1146/annurev.en.25.010180.002311) - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

