Novel patterns of expression and recruitment of new genes on the t-haplotype, a mouse selfish chromosome
- PMID: 35135349
- PMCID: PMC8826135
- DOI: 10.1098/rspb.2021.1985
Novel patterns of expression and recruitment of new genes on the t-haplotype, a mouse selfish chromosome
Abstract
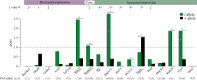
The t-haplotype of mice is a classical model for autosomal transmission distortion. A largely non-recombining variant of the proximal region of chromosome 17, it is transmitted to more than 90% of the progeny of heterozygous males through the disabling of sperm carrying a standard chromosome. While extensive genetic and functional work has shed light on individual genes involved in drive, much less is known about the evolution and function of the rest of its hundreds of genes. Here, we characterize the sequence and expression of dozens of t-specific transcripts and of their chromosome 17 homologues. Many genes showed reduced expression of the t-allele, but an equal number of genes showed increased expression of their t-copy, consistent with increased activity or a newly evolved function. Genes on the t-haplotype had a significantly higher non-synonymous substitution rate than their homologues on the standard chromosome, with several genes harbouring dN/dS ratios above 1. Finally, the t-haplotype has acquired at least two genes from other chromosomes, which show high and tissue-specific expression. These results provide a first overview of the gene content of this selfish element, and support a more dynamic evolutionary scenario than expected of a large genomic region with almost no recombination.
Keywords: gene gain; neofunctionalization; transmission distortion.
Conflict of interest statement
We declare we have no competing interests.
Figures



References
-
- Oestergren G. 1945. Parasitic nature of extra fragment chromosomes. Bot. Not. 2, 157-163.
-
- Sandler L, Novitski E. 1957. Meiotic drive as an evolutionary force. Am. Nat. 91, 105-110. (10.1086/281969) - DOI
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources

