Genome editing techniques in plants: a comprehensive review and future prospects toward zero hunger
- PMID: 35135438
- PMCID: PMC9208631
- DOI: 10.1080/21645698.2021.2021724
Genome editing techniques in plants: a comprehensive review and future prospects toward zero hunger
Abstract
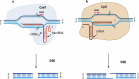
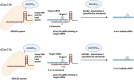
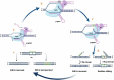
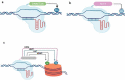
Promoting sustainable agriculture and improving nutrition are the main united nation sustainable development goals by 2030. New technologies are required to achieve zero hunger, and genome editing technology is the most promising one. In the last decade, genome editing (GE) using the CRISPR/Cas system has attracted researchers as a safer and easy tool for genome editing in several living organisms. GE has revolutionized the field of agriculture by improving biotic and abiotic stresses and yield improvement. GE technologies were developed fast lately to avoid the obstacles that face GM crops. GE technology, depending on site directed nuclease (SDN), is divided into three categories according to the modification methods. Developing transgenic-free edited plants without introducing foreign DNA meet the acceptance and regulatory ratification of several countries. There are several ongoing efforts from different countries that are rapidly expanding to adopt the current technological innovations. This review summarizes the different GE technologies and their application as a way to help in ending hunger.
Keywords: CRISPR/Cas; base editing; crop improvenet; food security; prime editing.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
