NK cell frequencies, function and correlates to vaccine outcome in BNT162b2 mRNA anti-SARS-CoV-2 vaccinated healthy and immunocompromised individuals
- PMID: 35135470
- PMCID: PMC8822735
- DOI: 10.1186/s10020-022-00443-2
NK cell frequencies, function and correlates to vaccine outcome in BNT162b2 mRNA anti-SARS-CoV-2 vaccinated healthy and immunocompromised individuals
Abstract
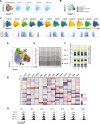
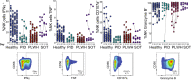
Adaptive immune responses have been studied extensively in the course of mRNA vaccination against COVID-19. Considerably fewer studies have assessed the effects on innate immune cells. Here, we characterized NK cells in healthy individuals and immunocompromised patients in the course of an anti-SARS-CoV-2 BNT162b2 mRNA prospective, open-label clinical vaccine trial. See trial registration description in notes. Results revealed preserved NK cell numbers, frequencies, subsets, phenotypes, and function as assessed through consecutive peripheral blood samplings at 0, 10, 21, and 35 days following vaccination. A positive correlation was observed between the frequency of NKG2C+ NK cells at baseline (Day 0) and anti-SARS-CoV-2 Ab titers following BNT162b2 mRNA vaccination at Day 35. The present results provide basic insights in regards to NK cells in the context of mRNA vaccination, and have relevance for future mRNA-based vaccinations against COVID-19, other viral infections, and cancer.Trial registration: The current study is based on clinical material from the COVAXID open-label, non-randomized prospective clinical trial registered at EudraCT and clinicaltrials.gov (no. 2021-000175-37). Description: https://clinicaltrials.gov/ct2/show/NCT04780659?term=2021-000175-37&draw=2&rank=1 .
Keywords: Anti-SARS-CoV-2 antibodies; BNT162b2 mRNA vaccine; COVID-19; Clinical trial; Innate immunity; NK cells; NKG2C; SARS-CoV-2; mRNA vaccine.
© 2022. The Author(s).
Conflict of interest statement
H.G.L. has served on the UK-CIC COVID-19 Oversight Committee, and leads the Karolinska Institutet COVID-19 vaccine group. H.G.L. is the Director of the NextGenNK Competence Center supported by Sweden’s Innovation Agency. Together with E.A., he is the founder and board member of XNK Therapeutics and Vycellix Inc. None of the other authors have declared any conflicts of interest in relation to the present study.
Figures


References
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

