DOT1L Is a Novel Cancer Stem Cell Target for Triple-Negative Breast Cancer
- PMID: 35135840
- PMCID: PMC9365344
- DOI: 10.1158/1078-0432.CCR-21-1299
DOT1L Is a Novel Cancer Stem Cell Target for Triple-Negative Breast Cancer
Abstract
Purpose: Although chemotherapies kill most cancer cells, stem cell-enriched survivors seed metastasis, particularly in triple-negative breast cancers (TNBC). TNBCs arise from and are enriched for tumor stem cells. Here, we tested if inhibition of DOT1L, an epigenetic regulator of normal tissue stem/progenitor populations, would target TNBC stem cells.
Experimental design: Effects of DOT1L inhibition by EPZ-5676 on stem cell properties were tested in three TNBC lines and four patient-derived xenograft (PDX) models and in isolated cancer stem cell (CSC)-enriched ALDH1+ and ALDH1- populations. RNA sequencing compared DOT1L regulated pathways in ALDH1+ and ALDH1- cells. To test if EPZ-5676 decreases CSC in vivo, limiting dilution assays of EPZ-5676/vehicle pretreated ALDH1+ and ALDH1- cells were performed. Tumor latency, growth, and metastasis were evaluated. Antitumor activity was also tested in TNBC PDX and PDX-derived organoids.
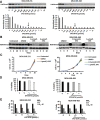
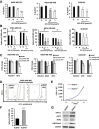
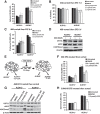
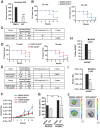
Results: ALDH1+ TNBC cells exhibit higher DOT1L and H3K79me2 than ALDH1-. DOT1L maintains MYC expression and self-renewal in ALDH1+ cells. Global profiling revealed that DOT1L governs oxidative phosphorylation, cMyc targets, DNA damage response, and WNT activation in ALDH1+ but not in ALDH1- cells. EPZ-5676 reduced tumorspheres and ALDH1+ cells in vitro and decreased tumor-initiating stem cells and metastasis in xenografts generated from ALDH1+ but not ALDH1- populations in vivo. EPZ-5676 significantly reduced growth in vivo of one of two TNBC PDX tested and decreased clonogenic 3D growth of two other PDX-derived organoid cultures.
Conclusions: DOT1L emerges as a key CSC regulator in TNBC. Present data support further clinical investigation of DOT1L inhibitors to target stem cell-enriched TNBC.
©2022 The Authors; Published by the American Association for Cancer Research.
Figures


References
-
- Tannock IF, Hill RP, Carey RW. The basic science of oncology. Plast Reconstr Surg 1989;83:920.
-
- Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: role of ATP–dependent transporters. Nat Rev Cancer 2002;2:48–58. - PubMed
-
- Vitale I, Manic G, De Maria R, Kroemer G, Galluzzi L. DNA damage in stem cells. Mol Cell 2017;66:306–19. - PubMed
-
- Wulf GG, Wang RY, Kuehnle I, Weidner D, Marini F, Brenner MK, et al. . A leukemic stem cell with intrinsic drug efflux capacity in acute myeloid leukemia. Blood 2001;98:1166–73. - PubMed
-
- Frank NY, Margaryan A, Huang Y, Schatton T, Waaga-Gasser AM, Gasser M, et al. . ABCB5-mediated doxorubicin transport and chemoresistance in human malignant melanoma. Cancer Res 2005;65:4320–33. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

