Genomic epidemiology of SARS-CoV-2 under an elimination strategy in Hong Kong
- PMID: 35136039
- PMCID: PMC8825829
- DOI: 10.1038/s41467-022-28420-7
Genomic epidemiology of SARS-CoV-2 under an elimination strategy in Hong Kong
Abstract
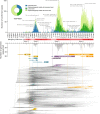
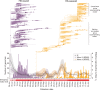
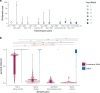
Hong Kong employed a strategy of intermittent public health and social measures alongside increasingly stringent travel regulations to eliminate domestic SARS-CoV-2 transmission. By analyzing 1899 genome sequences (>18% of confirmed cases) from 23-January-2020 to 26-January-2021, we reveal the effects of fluctuating control measures on the evolution and epidemiology of SARS-CoV-2 lineages in Hong Kong. Despite numerous importations, only three introductions were responsible for 90% of locally-acquired cases. Community outbreaks were caused by novel introductions rather than a resurgence of circulating strains. Thus, local outbreak prevention requires strong border control and community surveillance, especially during periods of less stringent social restriction. Non-adherence to prolonged preventative measures may explain sustained local transmission observed during wave four in late 2020 and early 2021. We also found that, due to a tight transmission bottleneck, transmission of low-frequency single nucleotide variants between hosts is rare.
© 2022. The Author(s).
Conflict of interest statement
B.J.C. has consulted for Roche, Sanofi Pasteur, GSK, AstraZeneca, and Moderna. The authors declare no other competing interests.
Figures

Update of
-
SARS-CoV-2 under an elimination strategy in Hong Kong.medRxiv [Preprint]. 2021 Jun 23:2021.06.19.21259169. doi: 10.1101/2021.06.19.21259169. medRxiv. 2021. Update in: Nat Commun. 2022 Feb 8;13(1):736. doi: 10.1038/s41467-022-28420-7. PMID: 34189537 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Grants and funding
- U01 AI151810/AI/NIAID NIH HHS/United States
- HHSN272201400006C/AI/NIAID NIH HHS/United States
- 75N93021C00016/AI/NIAID NIH HHS/United States
- COVID190118/Food and Health Bureau of the Government of the Hong Kong Special Administrative Region | Health and Medical Research Fund (HMRF)
- C7123-20G/Research Grants Council, University Grants Committee (RGC, UGC)
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

