The biogeography of infection revisited
- PMID: 35136217
- PMCID: PMC9357866
- DOI: 10.1038/s41579-022-00683-3
The biogeography of infection revisited
Abstract
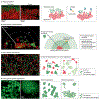
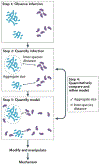
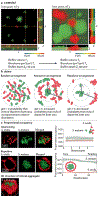
Many microbial communities, including those involved in chronic human infections, are patterned at the micron scale. In this Review, we summarize recent work that has defined the spatial arrangement of microorganisms in infection and begun to demonstrate how changes in spatial patterning correlate with disease. Advances in microscopy have refined our understanding of microbial micron-scale biogeography in samples from humans. These findings then serve as a benchmark for studying the role of spatial patterning in preclinical models, which provide experimental versatility to investigate the interplay between biogeography and pathogenesis. Experimentation using preclinical models has begun to show how spatial patterning influences the interactions between cells, their ability to coexist, their virulence and their recalcitrance to treatment. Future work to study the role of biogeography in infection and the functional biogeography of microorganisms will further refine our understanding of the interplay of spatial patterning, pathogen virulence and disease outcomes.
© 2022. Springer Nature Limited.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures
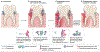

References
-
- Hall-Stoodley L, Costerton JW & Stoodley P Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol 2, 95–108 (2004). - PubMed
-
- Flemming HC & Wuertz S Bacteria and archaea on Earth and their abundance in biofilms. Nat. Rev. Microbiol 17, 247–260 (2019). - PubMed
-
- Azimi S, Klementiev AD, Whiteley M & Diggle SP Bacterial quorum sensing during infection. Annu. Rev. Microbiol 74, 201–219 (2020). - PubMed
-
- Martiny JB et al. Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol 4, 102–112 (2006). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

