Loop-Mediated Isothermal Amplification Detection of SARS-CoV-2 and Myriad Other Applications
- PMID: 35136384
- PMCID: PMC8802757
- DOI: 10.7171/jbt.21-3203-017
Loop-Mediated Isothermal Amplification Detection of SARS-CoV-2 and Myriad Other Applications
Abstract
As the second year of the COVID-19 pandemic begins, it remains clear that a massive increase in the ability to test for SARS-CoV-2 infections in a myriad of settings is critical to controlling the pandemic and to preparing for future outbreaks. The current gold standard for molecular diagnostics is the polymerase chain reaction (PCR), but the extraordinary and unmet demand for testing in a variety of environments means that both complementary and supplementary testing solutions are still needed. This review highlights the role that loop-mediated isothermal amplification (LAMP) has had in filling this global testing need, providing a faster and easier means of testing, and what it can do for future applications, pathogens, and the preparation for future outbreaks. This review describes the current state of the art for research of LAMP-based SARS-CoV-2 testing, as well as its implications for other pathogens and testing. The authors represent the global LAMP (gLAMP) Consortium, an international research collective, which has regularly met to share their experiences on LAMP deployment and best practices; sections are devoted to all aspects of LAMP testing, including preanalytic sample processing, target amplification, and amplicon detection, then the hardware and software required for deployment are discussed, and finally, a summary of the current regulatory landscape is provided. Included as well are a series of first-person accounts of LAMP method development and deployment. The final discussion section provides the reader with a distillation of the most validated testing methods and their paths to implementation. This review also aims to provide practical information and insight for a range of audiences: for a research audience, to help accelerate research through sharing of best practices; for an implementation audience, to help get testing up and running quickly; and for a public health, clinical, and policy audience, to help convey the breadth of the effect that LAMP methods have to offer.
© 2021 ABRF.
Figures
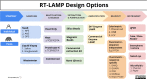
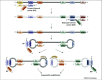


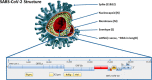
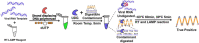
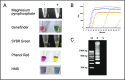


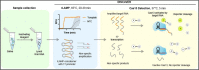
References
-
- Agrawal S, Fanton A, Chandrasekaran S, et al. Rapid, point-of-care molecular diagnostics with Cas13. medRxiv . 2020. Posted online April 5. and in revised form 2021. - DOI
-
- Aidelberg G, Aronoff R. Freeze-drying (lyophilization) of CoronaDetective tubes V.1. 2020. dx.doi.org/10.17504/protocols.io.bk44kyyw Available at: https://www.protocols.io/view/freeze-drying-lyophilization-of-coronadete... Posted September 8. - DOI
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
